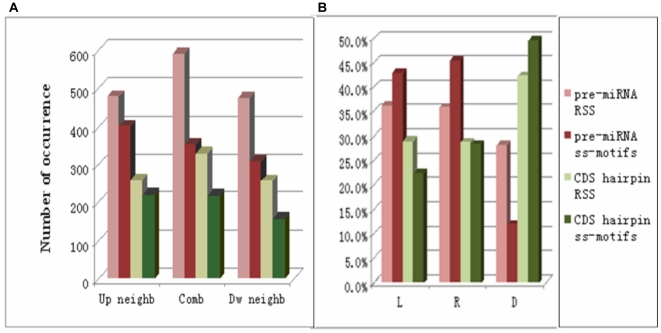
Figure 3. Sequence-structure motif characteristics.
Light hues (pink, light green) indicates the positive and negative randomly selected sequences (RSS). Darker hues (red, green) indicates the actual ss-motifs derived from the positive (pre-miRNA) and negative (CDS hairpin) training sets. A. Combinations of nucleotide and structural information in the ss-motifs. The figure shows occurrence of structural information relative to positions with specific nucleotide information. “Comb” denotes occurrences of specific nucleotide and structural information combined at the same position (e.g., “AL”, “CD”, etc). “Up neighb” and “Dw neighb” denote occurrences of specific nucleotide notation combined with specific structural notations at the nearest upstream or downstream neighbouring position (e.g., “NLAS” etc, and “ASNL” etc), respectively. B. Distribution of “L”, “R” and “D” denotes “left”, “right” and absence of (notation of) intramolecular interactions, respectively.