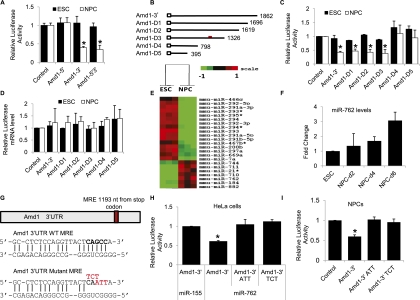
Figure 5.
Amd1 translational down-regulation is mediated by miR-762. (A) A bar chart showing the effect on luciferase activity of the Amd1 5′ UTR, 3′ UTR, or 5′ and 3′ UTRs in ESCs and NPCs. Luciferase values are normalized to cotransfected Renilla luciferase and empty vector. (B) Diagram showing the deletion constructs made to map the Amd1 3′ UTR cis-acting elements. The numbers refer to the length of the 3′ UTR remaining in each construct. The red mark indicates the location of the miR-762 target site. (C) Relative luciferase activities for the constructs shown in B after transfection into ESCs and NPCs. Values are normalized to firefly luciferase and empty vector. (D) qRT–PCR analysis of the relative luciferase mRNA levels after transfection into ESCs and NPCs. The Renilla luciferase is normalized to the firefly luciferase mRNA levels. (E) Heat map showing differentially expressed miRNAs from ESCs and NPCs. (F) qRT–PCR showing up-regulation of miR-762 in NPCs 2, 4, and 6 d after differentiation. (G) Predicted miR-762 miRNA response element (MRE) sequences within the Amd1 3′ UTR and its mutant counterpart. (H) Relative luciferase activity of PsiCHECK-2 constructs carrying the Amd1 wild-type 3′ UTR or Amd1 mutant 3′ UTR cotransfected into HeLa cells with miR-762. Values are normalized to control luciferase and empty vectors. (I) Relative luciferase activity of PsiCHECK-2 constructs carrying the Amd1 wild-type 3′ UTR or Amd1 mutant 3′ UTR cotransfected into ESCs and NPCs. Values are normalized to control luciferase and empty vector. Values are means ± SD. (*) P < 0.05.