Figure 3.
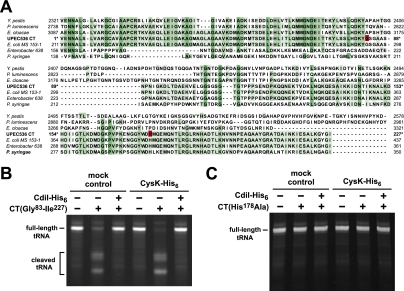
CdiA-CT is an intrinsic tRNase. (A) Multiple sequence alignment suggests that UPEC536 CdiA-CT is comprised of two domains. Homologous CdiA-CT regions from various proteobacteria were aligned with UPEC536 CdiA-CT using ClustalW. The N-terminal region of UPEC536 CdiA-CT (Val 1–Tyr 82) shares sequence identity with the corresponding regions in CdiA proteins from Y. pestis (Uniprot Q74T84), P. luminescens (Q7MB60), and E. cloacae (D5CBA0). The C-terminal region (Asn 89–Ile 227) is homologous to CdiA-related proteins from E. coli MS 153-1 (E6AEZ0), Enterobacter sp. 638 (A4W4W8), and Pseudomonas syringae (F3FGM0). UPEC536 CdiA-CT residues Gly 83 and His 178 (numbered with respect to Val 1 of the VENN motif) are highlighted in red. The alignment was rendered with 50% conservation visibility using Jalview 2.7. (B) The C-terminal domain of CdiA-CT has intrinsic tRNase activity. GITC-phenol-extracted tRNA was treated with the C-terminal domain of CdiA-CT (residues Gly 83–Ile 227) and CdiI-His6 in the presence and absence (mock control) of CysK-His6, and reactions were analyzed by denaturing gel electrophoresis. The migration positions of full-length and cleaved tRNAs are indicated. (C) The His178Ala missense mutation ablates CdiA-CT tRNase activity. GITC-phenol-extracted tRNA was treated with purified CdiA-CT(H178A) and CdiI-His6 in the presence and absence (mock control) of CysK-His6. Reactions were analyzed by denaturing gel electrophoresis.