Abstract
Actin forms a double-stranded filament, and the majority of actin filaments in the cell undergo the dynamic process of polymerization and depolymerization at both ends. Actin dynamics plays numerous important roles in eukaryotic cells. In order to understand actin dynamics, structural elucidation of the actin filament ends is particularly important because polymerization and depolymerization occurs only at the ends. We have developed original image analysis procedures to determine the structures of the actin filament ends from cryo-electron micrographs, and two structures have been determined. The structures revealed that the actin filament takes advantage of its double-stranded form to regulate its dynamics at both ends by a surprisingly simple mechanism.
Keywords: actin, structure, dynamics, cyro-electron microscopy, image analysis, cytoskeleton, end of the actin filament
Actin is one of the most abundant proteins in eukaryotic cells and forms a double-stranded filament which is involved in various kinds of cellular functions including cell adhesion, cell motility, cell division, cytoskeletal arrangement and muscle contraction. In most cases, the actin filament is dynamic through depolymerization and polymerization at the both ends. The actin subunits in a stress fiber are replaced by polymerization and depolymerization in several minutes.1 The actin dynamics in lamellipodia and philopodia is more typical. The polymerization at one specific end (the barbed end) and the depolymerization at the other end (the pointed end) push the cell membrane outward.2
To understand the regulatory mechanisms of actin dynamics, the determination of the actin filament end structures is crucial because depolymerization and polymerization occur only at the ends. We have developed a method to determine the end structure by cryo-electron microscopy and image analysis procedures.3 The two structures that were determined, the actin-Capping Protein (CP) complex4 and the bare pointed end of the actin filament,5 revealed unknown regulatory mechanisms of the actin dynamics. In these mechanisms, the actin filament takes advantage of its own double-stranded form in three different ways.
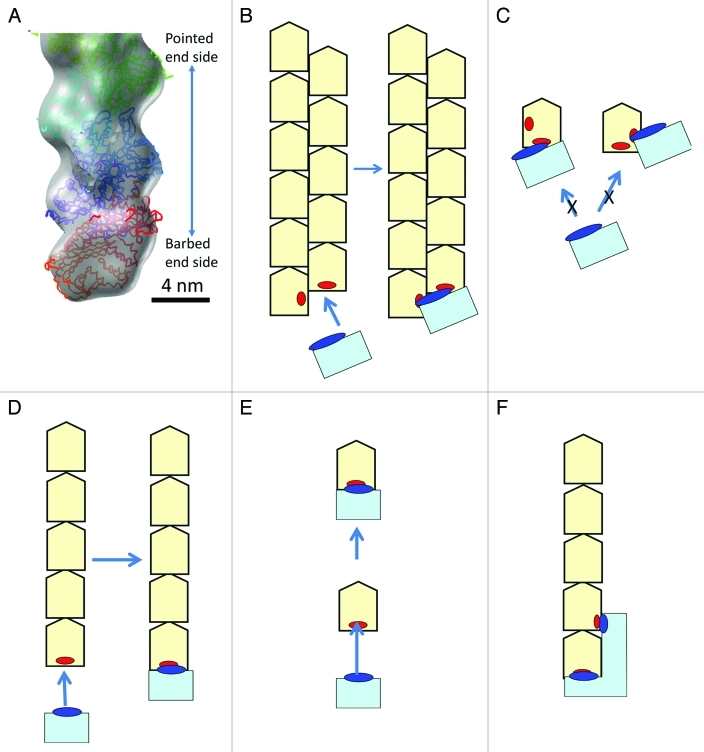
1. The double-stranded form is required for end-binding proteins to recognize and bind, and these proteins do not recognize actin monomers (Fig. 1). Many end binding proteins such as CP, formin, tropomodulin and spire,6-8 regulate actin dynamics because these proteins are most effective at binding to the filament ends where the polymerization and depolymerization occur. Therefore, recognition of the end by an end binding protein is important. The actin-CP complex structure represents a simple and sophisticated manner of the recognition process (Fig. 1A-C). We believe many end binding proteins recognize the target end by a similar manner, simultaneously binding to two regions which are exposed only at the target end on two subunits located on different strands. When the filament is single-stranded, the specific recognition of the target end is more difficult (Fig. 1D-F).
Figure 1.
The double-stranded form is useful for end binding proteins to recognize the target end. A-C: The binding mechanism of the Capping Protein.4 A: A three dimensional map of the actin-CP complex with fitted atomic models of CP (in red and orange) and actin molecules (in purple, blue, cyan and green). CP binds to only the barbed end, neither to the side of the filament, nor the pointed end, nor the actin monomer. B: A schematic illustration of CP (in cyan) binding to the barbed end. The major binding site on CP, illustrated as a blue ellipse, binds to the two end subunits on the two strands simultaneously. The binding sites on the actin filament, illustrated as red ellipses, are exposed only at the barbed end, neither at the pointed end nor the side of the filament, thereby illustrating how CP recognizes the barbed end. C: CP does not bind to actin monomers. CP requires two binding sites on two different strands with proper relative positioning for tight binding. CP can bind to only one binding site on the actin monomer even though one actin monomer has the two binding sites present (red ellipses). Consequently, the binding is significantly weaker than at the barbed end. D-F: Schematic illustrations of a putative model on how an end binding protein (in cyan) recognizes the end when the filament is single-stranded. D,E: Even when the end binding protein (the binding site is presented in blue) recognizes only the one end subunit (the binding site is presented in red), it can recognize the target end of the filament when the binding site on the filament is exposed only at the target end (D). However, it also binds to the monomer (E). To prevent binding to the monomer, the end binding protein (in cyan) must recognize two sites on the two subunits at the end including a site which is only exposed at the target end (F). The interaction between the end filament requires a much larger protein than with the double-stranded filament interaction model presented in (C).
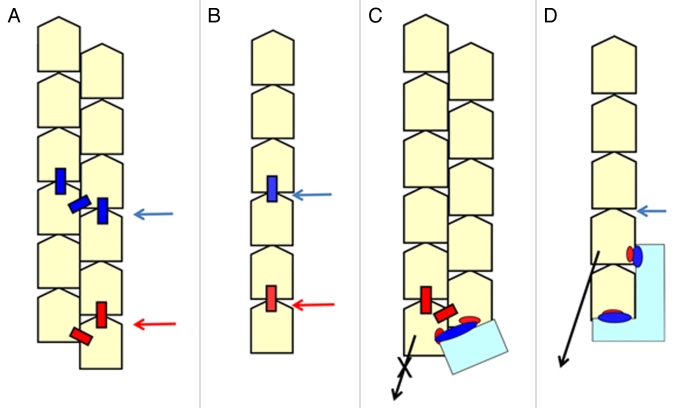
2: End binding proteins can regulate the stability of the whole filament (Fig. 2) when the filament has a double-stranded form. The actin filament in muscle is very stable, whereas it is truly dynamic in the lamellipodia where the turnover of the actin filament is less than one minute.9 The actin filament must cover a wide stability range and regulation of the stability by end binding proteins is essential.
Figure 2.
End binding proteins can regulate the stability of the whole filament when the filament is double-stranded. A: The depolymerization (dissociation) rate is much faster than the severing rate when the filament is double-stranded. This is because it is necessary to cut three bonds (the blue boxes indicated by a blue arrowhead) between the subunits at the same time to sever the filament, whereas cutting two bonds (the red boxes indicated by a red arrowhead) is sufficient for the end subunit to dissociate from the end. B: The depolymerization rate is similar to the severing rate when the filament is single-stranded, because the bond to cut for severing (the blue box) and depolymerizing (the red box) is identical. C: When CP (cyan) binds to the filament, the dissociation of the end subunit is prevented because an extra bond between CP and the end subunit must be cut in addition to the original two bonds (A, red boxes) for dissociation to occur from the end. Since the depolymerization rate is much faster than the severing rate, it is possible to increase the stability of the whole filament just by preventing the end subunit from dissociating with the double-stranded form. D: When the filament is single-stranded, the severing and depolymerizing rates are similar. Even the end binding protein bound to the two end subunits simultaneously (cyan, Figure 1F) cannot prevent the filament from severing where the binding protein does not bind (a blue arrowhead, for example) and the end binding protein will dissociate from the filament because of this severing event. Therefore, it is impossible to stabilize the whole filament by end binding proteins.
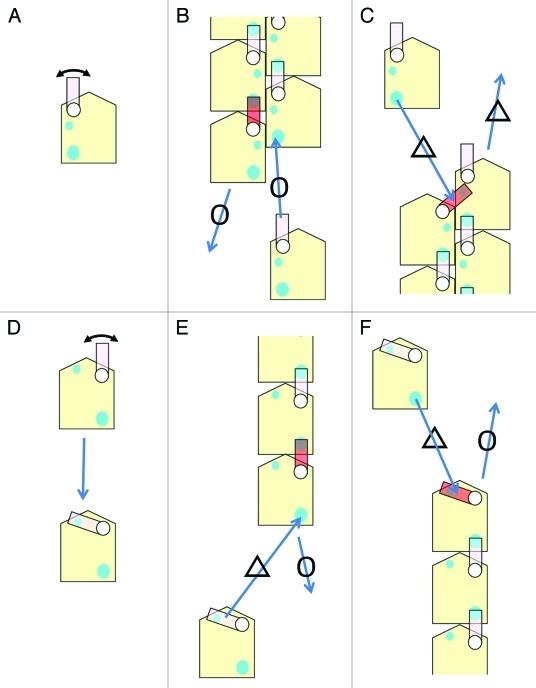
3: The double-stranded form plays an important role in defining the polarity of the dynamics (Fig. 3). One end, the pointed end, is much slower in polymerization and depolymerization than the barbed end, and these differences determine the direction of the “treadmilling” movement.5 We believe that this feature also defines the origin of the direction of the dynamics of the actin filament in a cell; where polymerization occurs at the barbed end and the depolymerization occurs at the pointed end.5
Figure 3.
The double-stranded form is important in defining the polarity of dynamics. A simplified model to explain the difference in the dynamics between the two ends is presented. A more realistic and complicated model was proposed previously;5 although the concept is the same. A: A simplified illustration of the actin monomer. The switching loop is the DNase-I binding loop in the simplified model. The actin monomer has two binding sites for the switching loop (cyan circles). One is a strong binding site (the large circle) and the other is a relatively weak binding site (the small circle). B: A model of the barbed end. The switching loop of the end subunit (in red) binds to the strong binding site on the actin subunit in the same strand. As a result, a strong binding site for an incoming actin monomer is available and the incoming monomer can easily bind to the end. C: A model of the pointed end. The switching loop of the adjacent subunit at the end (in red) binds to the weak binding site on the end subunit in the other strand. Consequently, the dissociation of the end subunit becomes more difficult at the pointed end because the extra binding by the switching loop (in red) prevents the end subunit from dissociating. When a new actin monomer comes in close proximity to bind to the strand, the switching loop (in red) that is already bound to a different site must first dissociate from the weaker site prior to interacting with strong binding site of the new monomer. As a result, the depolymerization and polymerization rates at the pointed end are slower than at the barbed end and represents the origin of the polarity of the dynamics of the actin filament. D-F: When the filament is single-stranded, it is difficult to define the polarity of the dynamics. If we assume a similar switching loop in the single-stranded filament, the weak binding site for the switching loop to bind at the pointed end must be located in the same strand because there is only one strand present. However, the switching loop in the monomer can also bind to the weak binding site in the same monomer (D). In this case, the polymerization at the pointed end (E) and at the barbed end (F) is inhibited to the same extent because of the switching loop binding to the weak binding site. For the dissociation from the ends, the situation cannot be different because the connection to be severed for dissociation to occur is identical between the two ends. Therefore, the rates of polymerization and depolymerization at the both ends must be similar when the filament is single-stranded.
In conclusion, our structures of the actin filament ends have revealed that the double-stranded form of the actin filament is essential in regulating actin dynamics in the cell, which is fundamental to all functions of the actin filament.
Glossary
Abbreviations:
- CP
Capping Protein
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/17137
References
- 1.Turnacioglu KK, Sanger JW, Sanger JM. Sites of monomeric actin incorporation in living PtK2 and REF-52 cells. Cell Motil Cytoskeleton. 1998;40:59–70. doi: 10.1002/(SICI)1097-0169(1998)40:1<59::AID-CM6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 2.Pantaloni D, Le Clainche C, Carlier MF. Mechanism of actin-based motility. Science. 2001;292:1502–6. doi: 10.1126/science.1059975. [DOI] [PubMed] [Google Scholar]
- 3.Narita A, Maeda Y. Molecular determination by electron microscopy of the actin filament end structure. J Mol Biol. 2007;365:480–501. doi: 10.1016/j.jmb.2006.06.056. [DOI] [PubMed] [Google Scholar]
- 4.Narita A, Takeda S, Yamashita A, Maeda Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 2006;25:5626–33. doi: 10.1038/sj.emboj.7601395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narita A, Oda T, Maeda Y. Structural basis for the slow dynamics of the actin filament pointed end. EMBO J. 2011;30:1230–7. doi: 10.1038/emboj.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zigmond SH. Beginning and ending an actin filament: control at the barbed end. Curr Top Dev Biol. 2004;63:145–88. doi: 10.1016/S0070-2153(04)63005-5. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Narita A, Hirayama T, Taki M, Iyoshi S, Yamamoto Y, et al. Human spire interacts with the barbed end of the actin filament. J Mol Biol. 2011;408:18–25. doi: 10.1016/j.jmb.2010.12.045. [DOI] [PubMed] [Google Scholar]
- 8.Fischer RS, Fowler VM. Tropomodulins: life at the slow end. Trends Cell Biol. 2003;13:593–601. doi: 10.1016/j.tcb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–6. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]