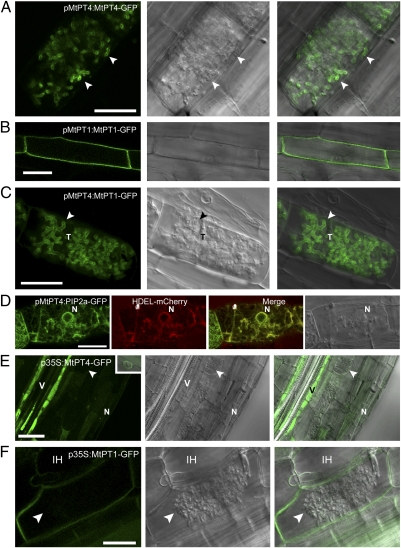
Fig. 1.
Protein localization in colonized cells is altered when genes are expressed from promoters that differ in activity relative to arbuscule development. Live-cell fluorescence and bright-field DIC images of longitudinal root sections of M. truncatula roots colonized with an AM fungus, G. versiforme. (A) pMtPT4:MtPT4-GFP localizes in the periarbuscular membrane (arrowhead) of colonized cortical cells. (B) pMtPT1:MtPT1-GFP localizes in the plasma membrane of root epidermal cells. (C) pMtPT4:MtPT1-GFP localizes in the periarbuscular membrane of colonized cortical cells and not in the plasma membrane. (D) pMtPT4:AtPIP2a-GFP does not localize in the periarbuscular or plasma membrane; instead it colocalizes with the HDEL-mCherry ER marker. (E) p35S:MtPT4-GFP accumulates mostly in the vacuole and ER, as shown by the presence of perinuclear signal (Inset), and not in the periarbuscular membrane (arrowhead). The signal is particularly strong in the vascular tissue. (F) p35S:MtPT1-GFP localizes in the plasma membrane and in the membrane that surrounds intracellular hyphae but not in the periarbuscular membrane. p35S:MtPT4-GFP includes the MtPT4 5′ UTR, whereas p35S:MtPT1-GFP contains the 35S 5′ UTR. For each construct, images are representative examples of at least 10 independent transgenic root systems; localization patterns were consistent among these independent experiments. Arrowheads indicate arbuscule branches. N, plant nucleus; T, arbuscule trunk; V, vascular tissue. (Scale bars: 20 μm.)