Abstract
Type I and type II interferons (IFNs) are cytokines that establish the cellular antiviral state through the induction of IFN-stimulated genes (ISGs). We sought to understand the basis of the antiviral activity induced by type I and II IFNs in relation to the functions of their ISGs. Based on gene expression studies, we systematically identified antiviral ISGs by performing blinded, functional screens on 288 type I and type II ISGs. We assessed and validated the antiviral activity of these ISGs against an RNA virus, vesicular stomatitis virus (VSV), and a DNA virus, murine gammaherpes virus (MHV-68). Overall, we identified 34 ISGs that elicited an antiviral effect on the replication of either one or both viruses. Fourteen ISGs have uncharacterized antiviral functions. We further defined ISGs that affect critical life-cycle processes in expression of VSV protein and MHV-68 immediate-early genes. Two previously undescribed antiviral ISGs, TAP1 and BMP2, were further validated. TAP1-deficient fibroblasts were more susceptible to VSV infection but less so to MHV-68 infection. On the other hand, exogenous BMP2 inhibits MHV-68 lytic growth but did not affect VSV growth. These results delineate common and distinct sets of type I and type II IFN-induced genes as well as identify unique ISGs that have either broad or specific antiviral effects on these viruses.
Keywords: interferon stimulated genes, antiviral effectors, murine gammaherpes virus 68
The immune system inhibits viral growth through expression of a diverse set of antiviral genes. Interferons (IFNs) are potent activators of these antiviral factors. Type I IFNs, which include IFNα, -β, and -ω, are rapidly activated during viral infection and considered “antiviral” partly because IFNα receptor-deficient mice are highly susceptible to viral infections (1). Type II IFN, which is represented only by IFNγ, also inhibits growth of viral and other pathogenic infections. Both types of IFN induce expression of IFN-stimulated genes (ISGs) that have a variety of functions ranging from direct inhibition of viral components to activation of other immune cell types.
Type I and type II IFNs have distinct physiological roles, but they both can activate the cellular antiviral response. Many cell types, particularly macrophages and dendritic cells, secrete type I IFN through the activation of intracellular and extracellular sensors of viral components. Type I IFN acts in paracrine and autocrine fashion to activate IFN receptors (IFNARs) present on most cell types. Upon receptor ligation, IFNAR is phosphorylated by the kinases JAK1 and TYK2, which recruit and phosphorylate STAT1 and STAT2 proteins. STAT1 can homodimerize or heterodimerize with STAT2 and translocate into the nucleus to activate specific target promoters (2). Type II IFN is largely secreted by T and natural killer cells and is induced predominantly by IL-12 and IL-18 (3). IFNγ receptor (IFNGR) activation leads to phosphorylation and homodimerization of STAT1 and target gene expression (3).
IFN-stimulated genes have diverse effects on different viruses and operate through distinct mechanisms. A comparison of ISGs that inhibit various RNA viruses shows that there are some that broadly inhibit RNA virus growth and others that are specific for particular viruses (4). We sought to identify and compare ISGs that inhibit growth of vesicular stomatitis virus (VSV), a negative-strand RNA virus, and murine gammaherpes virus 68 (MHV-68), a DNA virus. VSV is a neurotropic virus in the Rhabdoviridae family, which includes rabies virus. It infects most cell types and replicates lytically in the cytoplasm. MHV-68 is a member of the gammaherpes virus family that includes Kaposi sarcoma-associated herpes virus and Epstein–Barr virus. They can infect particular cell types and are thought to predominantly establish latency in B cells (5). These viruses are models for virology and several human-related diseases, yet a systematic identification of ISGs that inhibit these viruses has not been done.
In this study, we sought to compare IFNα and IFNγ induction of ISGs under an equivalent signaling input and systematically screened their effects on VSV and MHV-68 growth. We identified and validated known and unique ISGs that have broad and specific antiviral activity against these viruses.
Results
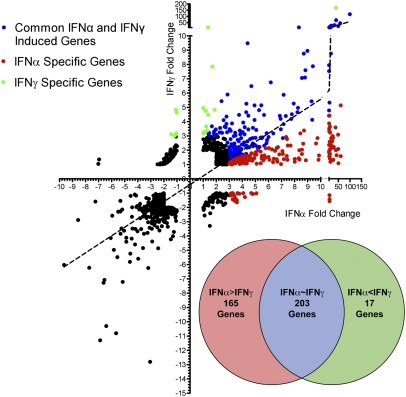
Both type I and type II IFNs broadly inhibit many types of viruses and elicit antiviral responses in various cell types. We hypothesized that IFNα and IFNγ can activate distinct and common ISGs that govern their antiviral effects. Because IFNα and IFNγ are structurally distinct cytokines that activate different receptors, we normalized IFNα and IFNγ concentrations based on phosphorylation of the shared and obligatory signaling factor STAT1 (6). Titration of both IFNs showed IFNγ phosphorylates STAT1 more than IFNα at equivalent units; for example, 1 U/mL of IFNγ phosphorylates STAT1 to an equivalent level as 62 U/mL of IFNα (Fig. S1 A and B). Based on this equivalent biological input, primary bone marrow-derived macrophages (BMMs) were stimulated with IFNα or IFNγ for 2.5 h in biological triplicates and processed for gene expression profiling by microarray (Fig. 1). Overall, IFNα and IFNγ regulated a common set of genes that had a significant correlation coefficient of 0.59. There were more ISGs induced by IFNα than IFNγ over untreated controls. Genes that were induced by IFNα threefold over IFNγ were classified as “IFNα-specific,” and genes that were induced by IFNγ threefold more than IFNα were classified as “IFNγ-specific.” Based on this categorization, 165 ISGs were IFNα-specific, 17 were IFNγ-specific, and 203 ISGs were commonly induced by both IFNs (Fig. 1, Inset and Table S1). These results suggest that under an equivalent signaling input, IFNα is a more efficient activator of gene expression than IFNγ.
Fig. 1.
Gene expression profile of BMMs treated for 2.5 h with IFNα and IFNγ at 62 U/mL and 1 U/mL, respectively. Axes represent fold change in response to IFNα or IFNγ over untreated cells. IFNα- and IFNγ-specific and commonly induced genes were categorized (see text) and are represented in a Venn diagram (Inset).
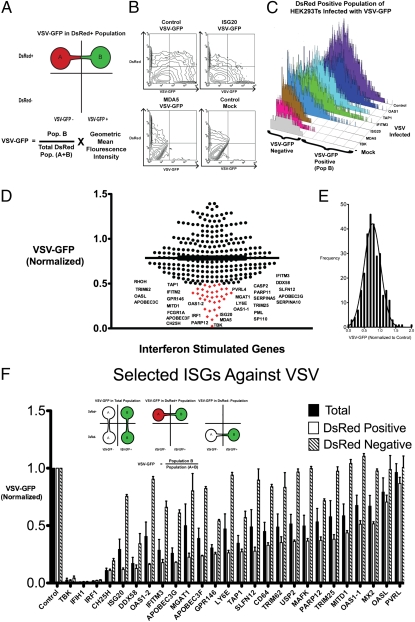
To further understand the basis of type I and type II antiviral activity, we next sought to identify IFNα- and IFNγ-induced ISGs that inhibit replication of VSV and MHV-68. The ISGs identified in Fig. 1 and Table S1 were used to generate a list of human ISGs based on homology for study in HEK293T cells. ISGs in the same family were included, and some genes were omitted because they were not found in the human genome. We obtained 288 genes in pCMV-driven expression constructs. First, we performed a blinded screen to identify ISGs that inhibit VSV coexpressing GFP (VSV-GFP) using a FACS-based approach. Individual ISGs in expression plasmids were cotransfected with red fluorescent construct (DsRed) in HEK293T cells. Empirical studies with a GFP construct and DsRed construct transfected at a 3:1 ratio show that >99% of DsRed-positive cells were also GFP-positive, which has been supported by other studies (7). Hence, DsRed-positive cells transfected under a 3:1 ratio of a gene of interest to DsRed would largely express the gene of interest. Using these conditions, HEK293T cells were cotransfected with individual ISGs and DsRed for 36 h and subsequently infected with VSV-GFP for 9 h and analyzed by FACS. Active viral replication was indirectly measured by GFP fluorescence in the cells. Tank-binding kinase 1 (TBK-1), which is a strong activator of IFN production, was used as positive control for the screen. The amount of infection was normalized to cells cotransfected with DsRed and a control gene, protein-tyrosine sulfotransferase 1 (TPST1), which had no effect on viral infection. As this screen was blinded, we sequenced the plasmids that inhibited viral growth to identify the gene.
For the initial large-scale screen, we measured VSV-GFP expression in the DsRed-positive population, which should be cells that highly coexpressed individual ISGs. We quantified VSV-GFP expression by the product of the percent VSV-GFP–positive, which should reflect the degree of viral infection across the population, and the geometric mean of the fluorescence intensity (MFI), which would reflect intensity of viral replication within a particular cell (Fig. 2 A and B). Next, we verified our approach with several known antiviral genes, such as IFITM3, ISG20, and OAS1, to serve as positive controls (Fig. 2C). Screening results demonstrated that expression of many of the ISGs inhibited VSV replication. Approximately 30 genes displayed greater than 50% inhibition of VSV-GFP expression, and the median VSV-GFP was 77% of cells transfected with control (Fig. 2 D and E). Known antiviral ISGs identified were IRF1, OAS1 variant 1 (OAS1-1), OAS1 variant 2 (OAS1-2), ISG20, IFIH (MDA5), IFITM3, TRIM25, RIG-I (DDX58), APOBEC3F, APOBEC3G, and OASL. ISGs that have uncharacterized antiviral activity were PARP11, PARP12, PVRL4, MGAT1, LY6E, CASP2, SERPAINA5, PML, SP110, SLFN12, SERPINA10, GPR146, MITD1, TAP1, RHOH, TRIM62, and FCGR1A (Table S2).
Fig. 2.
(A) Schematic of FACS-based assay to identify ISGs against VSV-GFP. HEK293T cells were transfected with individual ISGs with DsRed plasmid at a 3:1 ratio. DsRed-positive cells indicate cells that highly express the ISG. VSV growth (VSV-GFP) was calculated by the product of the percent GFP-positive cells and the geometric mean fluorescence intensity (MFI) of GFP in the DsRed-positive cells. (B) Contour maps of VSV-GFP in HEK293T cells transfected with the indicated ISGs and DsRed. (C) VSV-GFP was measured in the DsRed-positive population of selected ISGs transfected in HEK293T cells. TBK-1 was used as a positive control. (D) All 288 ISGs were screened by a FACS-based method. Each dot represents an ISG, and its effect on VSV-GFP expression is normalized to VSV-GFP in control transfected cells. ISGs that inhibited VSV-GFP expression over 50% are labeled and indicated in red. Values represent mean of duplicates. (E) Histogram of all ISGs and their effect on VSV-GFP normalized to VSV-GFP in control transfected cells. (F) Effect of individual ISGs on VSV-GFP expression in total population, DsRed-positive population, and DsRed-negative population was calculated. Values represent mean ± SEM from biological triplicates.
Twenty-four candidate genes that demonstrated high antiviral activity were chosen for further verification with biological triplicates. The amounts of VSV-GFP infection in total, DsRed-positive (DsRed+), and DsRed-negative (DsRed−) populations were measured for these ISGs and normalized to the control transfected cells of the respective populations (Fig. 2F). The DsRed-positive population should have high expression of the particular ISG that is cotransfected with DsRed, whereas the DsRed-negative population should have very low or no expression of the ISG. Comparison of both populations should indicate whether a particular ISG has an antiviral effect intrinsic to the cell expressing the gene or whether the ISG can confer protection in trans for cells expressing the gene. Genes that had the most pronounced inhibitory effect were IRF1, IFIH1 (MDA5), CH25H, and DDX58 (RIG-I). These genes inhibited VSV-GFP expression in all populations, suggesting that they amplify antiviral response to confer resistance on cells that do not express or express low levels of the ISG (DsRed-negative population), likely through induction of IFN (8, 9). On the other hand, ISGs such as ISG20, IFITM3, OAS1, MGAT1, GPR146, PARP12, LY6E, APOBEC proteins, TAP1, MX2, CD64, TRIM62, OASL, and others inhibited VSV-GFP expression in DsRed-positive cells but had little effect in DsRed-negative cells. These ISGs likely inhibit viral growth within the cell but not in surrounding cells. To validate this idea, we collected conditioned media from the supernatants of HEK293T cells transfected with IRF1, IFIH1, ISG20, and TBK and transferred them onto freshly plated HEK293T cells. The cells were infected with VSV after 4 h of treatment. Cells that were treated with MDA5, TBK, or IRF1 conditioned media were protective against VSV infection compared with control and ISG20-transfected conditioned media (Fig. S2). These results confirm that MDA5, TBK, and IRF1 can confer protection in trans to surrounding cells, whereas most other ISGs, such as ISG20, mediate their antiviral functions intrinsically. Taken together, this functional screen has identified known and unique antiviral ISGs against VSV (Table S3).
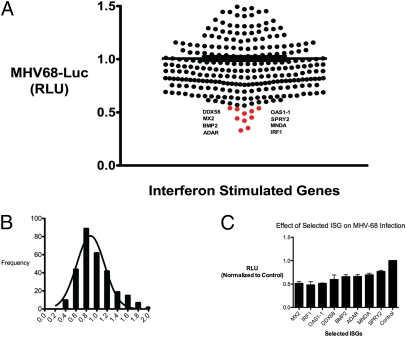
We next explored whether antiviral ISGs could be defined in similar or distinct sets based on their effect on growth of the DNA virus MHV-68. Individual ISGs were screened for their antiviral effect on MHV-68 coexpressing a luciferase reporter with the early-late gene, M3 (MHV-68-Luc) (10). HEK293T cells were transfected in quadruplicate with individual ISGs for 36 h and infected with MHV-68-Luc for 9 h, which was about the linear range for luciferase expression after infection. MHV-68 luciferase activity was measured and normalized to infected cells that expressed the control gene TPST1, which had no effect on MHV-68 replication (Fig. 3A and Table S4). The median reduction inhibition was about 10% (Fig. 3B). Eight genes that inhibited MHV-68-Luc production by ∼50% were verified in quadruplicate in independent experiments (Fig. 3C). We found that IRF1, MX2, DDX58, BMP2, SPRY2, MNDA, OAS1-1, and ADAR significantly inhibited MHV-68-Luc production (Table S5).
Fig. 3.
(A) Individual ISGs expressed in HEK293T cells and infected with MHV-68 at 0.25 MOI in quadruplicates. Cells were lysed at 9 hpi and luciferase activity was measured. Mean relative luminescence unit (RLU) are presented. (B) Histogram of all ISGs, and their effects on MHV-68-Luc were normalized to MHV-Luc in control transfected cells. (C) Selected ISGs from the screen were verified in two experiments in quadruplicate and normalized to control. Values are expressed as mean ± SEM.
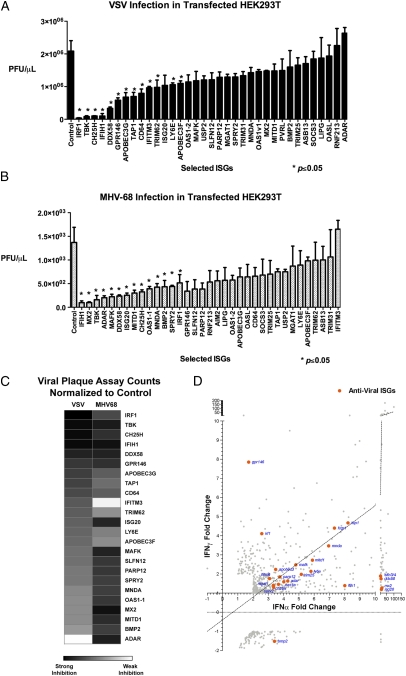
The screening approaches described thus far used indirect readouts using a marker, such as GFP or luciferase, coexpressed with a virally encoded protein. They are not a direct measure of infectious virions. Viral plaque assay is quantitative for live virions even though they are less sensitive than GFP or luciferase assays. Nonetheless, as a separate validation, we tested 34 selected ISGs from the previous two screens (Figs. 2F and 3C) and measured their effects on VSV or MHV-68 by plaque assay. Supernatants were collected at 9 h postinfection (hpi) for VSV and 24 hpi for MHV-68 and titered individually (Fig. 4 A and B). We considered ISGs that significantly inhibited viral growth (P ≤ 0.05 by Student's t test with unpaired, two-tailed hypothesis). IRF1, IFIH1, CH25H, ISG20, and DDX58 significantly inhibited growth of both viruses. ISGs that significantly antagonized VSV replication more strongly than MHV-68 were GPR146, APOBEC3G, APOBEC3F, TAP1, CD64, IFITM3, TRIM62, and LY6E. Conversely, ISGs that were more antiviral against MHV-68 than VSV were MNDA, BMP2, SPRY2, MAFK, OAS1-1, and ADAR (Fig. 4C). These results were significant and consistent in at least two independent experiments with biological triplicates. PARP12, GPR146, and SLFN12 demonstrated an inhibitory effect against the two viruses, but the results did not always meet the significance criteria (P < 0.05) in replicate experiments, indicating that they may have moderate inhibitory effects on viral replication. There were some ISGs such as PARP12, MITD1, ISG20, and MAFK that inhibited MHV-68 in plaque assays but did not affect luciferase expression, indicating that these ISGs may affect other viral life-cycle steps but not necessarily the activity of the MHV-68 M3 promoter of luciferase. Taken together, the plaque assay results provided additional validation of common and distinct sets of antiviral ISGs against VSV and MHV-68.
Fig. 4.
(A and B) The inhibitory effects of 34 ISGs selected from previous screens against VSV (A) and MHV-68 (B) were measured by plaque assay. Values represent mean ± SEM. Data represent 1 of 3 experiments. Asterisks indicate significant difference compared with control by Student's t test (P < 0.05). (C) Heat map showing the inhibitory effect of selected ISGs on VSV and MHV-68 based on plaque assay. (D) Antiviral ISGs (orange) was graphed with respect to their fold induction by IFNα and IFNγ in microarray analyses (Fig. 1).
The selected antiviral ISGs were compared with their respective type I and type II IFN-mediated gene expression based on the initial microarray study. Many antiviral ISGs were commonly induced by IFNα and IFNγ. IFNα induced most ISGs to higher levels than the correlation of all IFNα and IFNγ ISGs that were found to be antiviral (Fig. 4D, dotted line). Only IRF1 and GPR146 were induced by IFNγ higher than IFNα. These results provide an explanation for stronger inhibitory activity of IFNα against VSV and MHV-68 than IFNγ. The antiviral effect of IFNγ may be mediated by activation of fewer yet highly effective antiviral ISGs such as IRF1 or by activation of IFNα through an autocrine loop, as suggested in other studies (11–13).
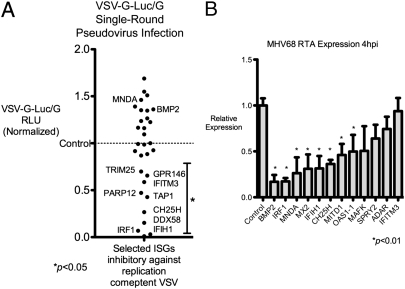
To better understand the effect of selected ISGs on inhibition of these viruses, we determined whether certain ISGs inhibited early critical life-cycle processes in VSV and MHV-68. As a negative-strand RNA virus, VSV undergoes primary transcription by its packaged polymerases to form positive-stranded mRNA; the process is independent of host protein synthesis after entry. Protein expression occurs in the sequential order N, P, M, G, and L, and is required for the switch from primary transcription to replication and downstream transcript amplification (14, 15). Hence, we tested whether the 30 most inhibitory ISGs against VSV from Fig. 4A could inhibit VSV protein expression. We used a VSV pseudovirus that has the receptor-binding G protein (VSV-G) replaced by the luciferase reporter (VSVΔG-Luc) enveloped inside VSV-G, called VSVΔG-Luc/G (16). This pseudovirus can undergo VSV-G–mediated entry but cannot produce its own VSV-G envelope, and hence is only capable of a single-round infection. Quantification of luciferase activity is indicative of viral life-cycle processes from entry to protein synthesis. MNDA and BMP2, which did not affect VSV replication, were used as negative controls. Of the 34 ISGs that inhibited VSV, only IRF1, CH25H, IFITM3, GPR146, TAP1, DDX58, PARP12, TRIM25, and IFIH1 inhibited VSVΔG-Luc expression at 8 hpi (Fig. 5A). Thus, these ISGs inhibited steps before viral protein expression. Other ISGs that antagonized VSV, such as OAS1, ISG20, TRIM62, APOBEC3G, and CD64, did not reduce VSVΔG-Luc activity, suggesting they inhibited subsequent steps in replication, secondary transcription, assembly, or budding. These results demonstrate antiviral ISGs that inhibit stages up to or after VSV protein expression.
Fig. 5.
(A) ISGs that were most inhibitory against VSV in Fig. 4A were transfected in HEK293T cells for 36 h in triplicate and infected with VSVΔG-Luc reporter within a VSV-G envelope. Cells were lysed 10 hpi and luciferase activity was measured. BMP2 and MNDA, which did not inhibit VSV replication, were used as negative controls. Mean RLU are presented; data are representative of two independent experiments. (B) Eleven ISGs that were most inhibitory against MHV-68 were expressed in HEK293T cells for 36 h and infected with 0.2 MOI of MHV-68. Expression of the immediate-early gene RTA was measured by quantitative PCR at 4 hpi. IFITM3, which has no inhibitory effect on MHV-68, was used as a negative control. Data are representative of three independent experiments.
For ISGs that inhibited replication of MHV-68, we determined whether critical early life-cycle processes were inhibited by the selected 11 ISGs in Fig. 4B (MNDA, IRF1, MX2, BMP2, MITD1, IFIH1, MAFK, CH25H, OAS1-1, ADAR, SPRY2). As a DNA virus, MHV-68 first expresses essential genes after entry into the cell independent of host protein synthesis, called immediate-early (IE) genes. The replication and transactivator protein RTA is an IE gene that activates subsequent MHV-68 gene expression and is required for lytic replication and reactivation from latency (17). Hence, we tested the effect of expression of selected ISGs on RTA expression in HEK293T cells. BMP2, MX2, IRF1, IFIH1, CH25H, MITD1, MNDA, and OAS1-1 significantly inhibited RTA expression at 4 hpi (P < 0.01) (Fig. 5B). IFITM3, which did not inhibit MHV-68 replication, served as a negative control. As a separate validation, MNDA, IRF1, BMP2, MX2, and MITD1 also inhibited another IE gene, ORF57, at 4 hpi with P < 0.01 (Fig. S3). SPRY2, ADAR, and MAFK did not inhibit RTA or ORF57 expression, indicating they might affect early or late MHV-68 gene expression or other late life-cycle processes. Taken together, these results delineate inhibitory ISGs against MHV-68 that affect stages up to or after expression of critical immediate-early genes.
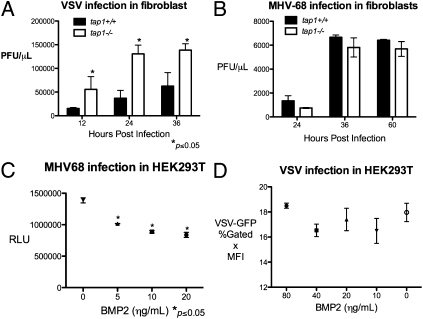
As a way to independently confirm our screening results, we sought to characterize two ISGs, TAP1 and BMP2, that have not been described as antiviral and had differential inhibitory effects on VSV and MHV-68 replication. Transporter associated with antigen processing 1 (TAP1) is well-described for its role in antigen presentation with MHC class I but has not been described to play a role in innate antiviral response. TAP1 expression significantly inhibited VSV replication but not MHV-68 replication. We hypothesized that deficiency in tap1 would adversely affect innate immune response against VSV. Hence, we infected tail-derived fibroblasts from tap1+/+ and tap1−/− mice with VSV and MHV-68. VSV growth in tap1−/− fibroblasts was significantly higher compared with tap1+/+ fibroblasts as assessed by plaque assay and FACS (Fig. 6A and Fig. S4A). There was no difference in MHV-68 replication by plaque assay or MHV-68-Luc expression (Fig. 6B and Fig. S4B). These results show that TAP1 is sufficient and may be a required factor for cellular immune response against VSV.
Fig. 6.
(A) tap1+/+ and tap1−/− tail-derived fibroblasts were infected with VSV at 0.1 MOI for the indicated time, and the supernatants were titered by plaque assay. Values represent mean ± SEM. (B) tap1+/+ and tap1−/− fibroblasts were infected the MHV-68 at 0.25 MOI, and the supernatants were collected at the indicated times and titered by plaque assay. Values represent mean ± SEM. (C) HEK293T were treated with hBMP2 at the indicated concentration for 12 h and infected with MHV-68-Luc at 0.25 MOI. Luciferase activity in the cell lysates was quantified at 9 hpi. Values represent mean ± SEM. RLU, relative luminescence unit. (D) HEK293T were treated with BMP2 at increasing concentrations for 12 h and infected with VSV at 0.01. VSV-GFP expression was measured by FACS at 9 hpi. Values represent mean ± SEM. MFI, geometric mean of fluorescence index.
Bone morphogenic protein 2 (BMP2) is well-studied in development, but its role in control of viral infection is unclear. Overexpression of bmp2 inhibited MHV-68-Luc expression and replication by 40% by plaque assay, but did not significantly affect VSV. Addition of the recombinant, active form of human human BMP2 (hBMP2) to HEK293T and murine pre-B-cells inhibited MHV-68-Luc expression in a dose-dependent manner (Fig. 6C and Fig. S4C). Exogenous addition of hBMP2 had no effect on VSV-GFP infection in HEK293T cells or B-cells (Fig. 6D and Fig. S4D). In addition, hBMP2 treatment of HEK293T inhibited RTA expression (Fig. S5). These results demonstrate BMP2 to be a sufficient inhibitory factor against MHV-68 but not VSV.
Discussion
In this study, we sought to understand the basis of the antiviral activity of type I and type II IFNs. IFNs induce a large group of ISGs, each of which is presumed to have important roles in innate immunity against different families of microorganisms. We performed functional screens of 288 type I and type II ISGs for their antiviral activity against VSV and MHV-68, which are RNA and DNA viruses, respectively. We found distinct and common sets of ISGs that inhibited VSV and MHV-68, including 14 ISGs that were not previously described as having antiviral activity. Of these newly identified antiviral ISGs, some inhibited critical early viral life-cycle processes in VSV protein expression and MHV-68 transcription. TAP1 and BMP2 were further functionally validated and shown to have specific, unique antiviral functions on these disparate viruses.
The large-scale functional screens identified more ISGs that inhibited VSV than MHV-68. This may be a reason that wild-type mice, which have a robust IFN response, are completely resistant to VSV but not to MHV-68 infection (1). IFNα is also a stronger inducer of antiviral ISGs than IFNγ, which is consistent with studies showing that type I IFN plays a dominant antiviral role. For example, ifnar−/− mice are very sensitive to VSV infection, whereas ifngr−/− mice are resistant (1). Immune-mediated inhibition of acute MHV-68 infection also requires type I but not type II IFNs (18, 19).
Interestingly, most antiviral ISGs inhibited these viruses moderately when expressed individually, suggesting that IFN-stimulated cells express a large group of ISGs that play a cumulative antiviral effect. Indeed, expression of various combinations of ISGs has an additive inhibitory effect on viruses (4). Expression of an array of antiviral effectors may be an effective method for the host to defend against the various viruses as well as counteract viral inhibition of any one or group of ISGs. Moreover, gene profiling suggests that many antiviral ISGs are induced only a few fold above basal levels (Fig. 4D). Expression of several ISGs at low levels may create less detrimental cellular changes while still achieving a global antiviral effect.
Many ISGs such as IRF1 and IFIH1 (MDA5) have broad antiviral activity, which can be attributed to amplification of IFN production. IRF1 is a well-studied transcription factor that induces IFN expression (20), and IFIH1 acts as an intracellular RNA receptor that activates IFN. Indeed, some studies support that this IFN amplification loop is a mechanism by which IFNγ mediates its antiviral effect. For example, IFNGR signaling is dependent on IFNAR activity (12), and STAT2, which is primarily activated by IFNAR, is required for IFNGR-mediated antiviral effect (11). Our data show that IFNγ is less efficient at activating most ISGs than IFNα, but induces IRF1 to higher levels than IFNα. IRF1 can directly induce ISGs as well as expression of type I IFN, implying that the antiviral activity of IFNγ may be mediated by the induction of IRF1 and type I IFN. In addition to positive feedback mechanisms, there may be other feedback mechanisms that may differentiate the gene expression programs and physiological effects of type I and II IFNs.
Several ISGs inhibited growth of VSV more than MHV-68, such as TAP1 IFITM3, GPR146, APOBEC proteins, CD64 (FCGR1A), TRIM62, and LY6E. Of these, GPR146, CH25H, PARP12, and TAP1 are unique antiviral proteins that inhibit VSV-G protein expression. It is likely that these ISGs inhibit steps before replication and secondary transcription, because sufficient protein translation is required before these steps.
To verify one of these genes, we showed that TAP1 is sufficient and required for inhibition of VSV in fibroblasts but not MHV-68, introducing a unique role of TAP1 in innate immune defense. This endoplasmic ATP-Binding Cassette (ABC) transporter facilitates transport of proteosomal-degraded proteins into the lumen onto MHC class I receptors. Interestingly, many viruses evade host immune response by inhibiting TAP1, which most studies attribute to be a method to block adaptive immune presentation to T cells. However, our work shows that there may be an antiviral function of TAP1 independent of its association with adaptive immune presentation. Its inhibitory effect on VSV-G-Luc expression suggests a potential role in posttranslational processing and degradation of viral proteins. A possible reason TAP1 did not play a significant role in inhibition of MHV-68 is that the viral mK3 protein of MHV-68 interacts with TAP1 and promotes its proteosomal degradation (21, 22).
ISGs that significantly inhibited MHV-68 more strongly than VSV include MX2, MNDA, SPRY2, BMP2, and ADAR1. BMP2 and MNDA are unique antiviral ISGs that strongly inhibited expression of the immediate-early gene RTA at 4 hpi. Expression of RTA directly affects expression of other MHV-68 genes required for lytic replication. We further showed that addition of recombinant BMP2 inhibits MHV-68 replication and RTA expression. One possible mechanism is that BMP2 can activate TGFβ-activating kinase 1 and subsequent activation of NF-κB, which prevents MHV-68 lytic growth (23, 24). This result introduces the possibility of unique IFN-inducible soluble factors that can inhibit growth of specific viruses.
Although more studies need to be done to elucidate the mechanisms of action and breadth of antiviral activity of the antiviral ISGs defined here, further understanding of these ISGs may provide future direction for antiviral therapies. Comprehensive understanding of ISG antiviral activity may introduce other avenues for targeted antiviral therapy that would bypass the need for IFN treatment as well as viral evasion strategies that inhibit IFN activation.
Materials and Methods
Cells and Reagents.
RAW and HEK293T cells were obtained from the American Type Culture Collection and grown in standard DMEM with 5% FBS, 1% penicillin/streptomycin (GIBCO). Glen Barber (University of Miami, Miami, FL) provided VSV-GFP. MHV-68 coexpressing luciferase reporter was provided by Ren Sun (University of California, Los Angeles, CA). Luciferase activity was measured using a firefly luciferase substrate kit (Promega). Human recombinant BMP2 was purchased from R&D Laboratories.
Bone marrow macrophages were harvested from 6- to 8-wk-old C57B/L6 mice (The Jackson Laboratory) and differentiated with 10 ng/mL of M-CSF in DMEM, 10% FBS for 7 d. On day 6 the media were replaced, and on day 7 the cells were stimulated with IFNα or IFNγ (PBL InterferonSource). The cells were treated for 2.5 h and harvested in TRIzol (Invitrogen). RNA was isolated by isopropanol precipitation for microarray analysis. For immunoblots, BMMs were treated with IFNα and IFNγ for 30 min and separated by SDS/PAGE as described previously (26) and blotted with phospho-STAT1 antibody (Cell Signaling). Primary antibodies were detected with anti-rabbit antisera conjugated to HRP (Santa Cruz Biotechnology) and visualized by chemiluminescence.
TAP1-deficient (Taptm1Arp) mice were purchased from The Jackson Laboratory. The tails of the mice were skinned and cultured in DMEM with 10% FBS, 1% (vol/vol) penicillin/streptomycin. Fibroblasts were harvested after 7–10 d. Murine pre-B Cells were derived from bone marrow as described previously (25).
Microarray and ISGs.
Microarrays were done on an Affymetrix 430.2 chip (University of California, Los Angeles Genotyping and Sequencing Core). Individual ISGs were provided by GeneCopoeia, Inc.
VSV Screening and Flow Cytometry.
HEK293T cells were plated in 12-well collagen-coated plates of 0.5 mg/mL rat tail collagen I (BD Biosciences) in PBS. Individual ISG expression plasmids were transfected with DsRed construct (Clontech) at a 3:1 ratio with FuGENE 6 (Roche) transfection reagent. After 36 h, the cells were infected with VSV-GFP at a 0.01 multiplicity of infection (MOI) and collected at 9 hpi in 2% (vol/vol) paraformaldehyde solution in PBS. FACS was done with standard compensation (FACSCaliber; BD Biosciences), and the data were analyzed using CellQuest (BD Biosciences).
MHV-68 Screening.
HEK293T cells were seeded in a 96-well plate overnight and transfected with individual ISG expression plasmids for 36 h. Cells were infected with MHV-68 at a 0.25 MOI for 1 h and lysed in 1× passive lysis buffer (Promega) at 9 hpi, and luciferase activity was measured.
Viral Plaque Assay.
Plaque assays were done on Vero cells in 12-well plates at 2 × 105 and 2 × 104 cells per well for VSV and MHV-68 plaque assays, respectively. Supernatants from infected cells were serially diluted and infected in Vero cells for 1 h. The cells were then covered with growth medium containing 0.6% (mass/vol) low-melting point agarose. Plaques were counted after 16 h or 6 d for VSV and MHV-68, respectively.
VSV-G–Pseudotyped VSV-G Luciferase.
VSV-G–pseudotyped VSV-G luciferase pseudovirus (VSVΔG-Luc/G) was a gift from Benhur Lee (University of California, Los Angeles, CA). The pseudovirus was generated by methods previously described (27) and concentrated by ultracentrifugation (>140,000 × g). The concentrations used to generate a linear range of luciferase signal were determined empirically.
MHV-68 Gene Expression Studies.
RNA was isolated from MHV-68–infected cells at the indicated times. cDNA was generated with iScript (Bio-Rad). Primers used for quantitative PCR were RTA-forward 5′-GATTCCCCTTCAGCCGATAAG-3′, RTA-reverse 5′-CAGACATTGTAGAAGTTCAGGTC-3′, ORF57-forward 5′-GACCAAATGATGGAAGGAC-3′, ORF57-reverse 5′-GCAGAGGAGAGTTGTGGAC-3′. PCR conditions used were 95 °C for 3 min followed by 45 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 30 s.
Software and Graphing.
Microarray analysis was performed using GeneSpring software (Agilent). All graphs were generated with GraphPad Prism. Heat maps were generated by using the web program Matrix2png (28).
Supplementary Material
Acknowledgments
We thank Dr. Ting-Ting Wu for her advice on MHV-68 gene expression studies and Dr. Edward Fritsch for editing the paper. This work was supported by National Institutes of Health (NIH)/National Cancer Institute Tumor Immunology Training Grant 5T32CA009120, NIH Grants R01 AI078389 and AI069120, Warsaw Fellowship, and the Medical Scientist Training Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35825).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114981109/-/DCSupplemental.
References
- 1.van den Broek MF, Müller U, Huang S, Zinkernagel RM, Aguet M. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol Rev. 1995;148(1):5–18. doi: 10.1111/j.1600-065x.1995.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 2.Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: An overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75(2):163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 4.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaño E, Kim I-J, Woodland DL, Blackman MA. γ-Herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J Exp Med. 2002;196:1363–1372. doi: 10.1084/jem.20020890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meraz MA, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 7.Xie ZL, et al. Co-transfection and tandem transfection of HEK293A cells for overexpression and RNAi experiments. Cell Biol Int. 2011;35(3):187–192. doi: 10.1042/CBI20100470. [DOI] [PubMed] [Google Scholar]
- 8.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto M, et al. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 10.Hwang S, et al. Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. J Virol. 2008;82:12498–12509. doi: 10.1128/JVI.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 12.Takaoka A, et al. Cross talk between interferon-γ and -α/β signaling components in caveolar membrane domains. Science. 2000;288:2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 13.Müller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee AK. Transcription and replication of rhabdoviruses. Microbiol Rev. 1987;51(1):66–87. doi: 10.1128/mr.51.1.66-87.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr JN, Whelan SP, Wertz GW. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim Biophys Acta. 2002;1577:337–353. doi: 10.1016/s0167-4781(02)00462-1. [DOI] [PubMed] [Google Scholar]
- 16.Negrete OA, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 17.Wu T-T, Usherwood EJ, Stewart JP, Nash AA, Sun R. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J Virol. 2000;74:3659–3667. doi: 10.1128/jvi.74.8.3659-3667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutia BM, Allen DJ, Dyson H, Nash AA. Type I interferons and IRF-1 play a critical role in the control of a gammaherpesvirus infection. Virology. 1999;261(2):173–179. doi: 10.1006/viro.1999.9834. [DOI] [PubMed] [Google Scholar]
- 19.Sarawar SR, et al. γ interferon is not essential for recovery from acute infection with murine gammaherpesvirus 68. J Virol. 1997;71:3916–3921. doi: 10.1128/jvi.71.5.3916-3921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi T, Takaoka A. The interferon-α/β system in antiviral responses: A multimodal machinery of gene regulation by the IRF family of transcription factors. Curr Opin Immunol. 2002;14(1):111–116. doi: 10.1016/s0952-7915(01)00305-3. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Lybarger L, Connors R, Harris MR, Hansen TH. Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex. J Virol. 2004;78:8673–8686. doi: 10.1128/JVI.78.16.8673-8686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boname JM, de Lima BD, Lehner PJ, Stevenson PG. Viral degradation of the MHC class I peptide loading complex. Immunity. 2004;20:305–317. doi: 10.1016/s1074-7613(04)00047-0. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi K, et al. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J. 1999;18:179–187. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown HJ, et al. NF-κB inhibits gammaherpesvirus lytic replication. J Virol. 2003;77:8532–8540. doi: 10.1128/JVI.77.15.8532-8540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherle PA, Dorshkind K, Witte ON. Clonal lymphoid progenitor cell lines expressing the BCR/ABL oncogene retain full differentiative function. Proc Natl Acad Sci USA. 1990;87:1908–1912. doi: 10.1073/pnas.87.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarnegar BJ, et al. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takada A, et al. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlidis P, Noble WS. Matrix2png: A utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.