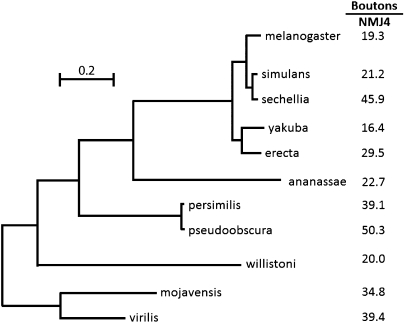
Fig. 4.
NMJ phenotypes among different species of Drosophila lack phylogenetic structure. The phylogenetic tree is derived from the complete genomic sequences of the species shown on the basis of rates of neutral substitutions at 12,000 sites (12). (Scale bar, number of substitutions per site.) The total evolutionary distance spanned by this phylogeny is >40 Myr with the distance from D. melanogaster to D. mojavensis being estimated at 40 Mya (4). Shown to the right of each species is the average number of boutons per NMJ4 as a representative quantitative measure of NMJ phenotype for that species (Table 1). Examination of the data reveals no obvious relationship between phylogenetic distance between the various species and their phenotypic similarity. The same is true if NMJ phenotypes are represented instead by number of synaptic branches.