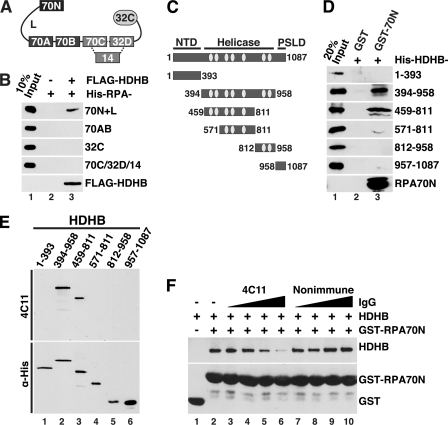
FIGURE 4.
Direct physical interaction of HDHB with RPA. A, modular domain architecture of RPA is shown. The three subunits RPA70, -32, and -14 interact through a three-helix bundle (dotted lines). Compact domains (filled rectangles, oligonucleotide/oligosaccharide binding-fold; filled oval, winged helix) are joined by flexible linkers (Lines) (44–48). The residues at the N and C termini of each domain are listed under “Experimental Procedures.” B, purified His-tagged RPA constructs captured on anti-FLAG antibody beads in the presence (+) or absence (−) of whole cell extract as in Fig. 3C were analyzed by Western blotting with anti-His (top four panels) or anti-FLAG. C, diagram of HDHB domains and truncation constructs. NTD, N-terminal domain; PSLD, phosphorylation regulated subcellular localization domain (20). The seven conserved helicase motifs I, Ia, and II-VI of superfamily I are indicated by light gray ovals. The first and last residue numbers of each construct are indicated. D, glutathione beads prebound to purified GST or His-GST-RPA70N were incubated with purified His-tagged HDHB truncation mutants. Proteins captured on the beads were analyzed by Western blotting with anti-His antibody. E, His-tagged HDHB truncation mutants (E) were separated by SDS-PAGE and visualized by Western blotting performed with HDHB monoclonal antibody 4C11 (top panel) or anti-His antibody (lower panel). F, glutathione beads prebound with GST (lane 1) or GST-RPA70N were incubated with purified HDHB (0.5 μg) in the absence (lanes 1 and 2) or presence of increasing amounts (0.3, 0.6, 1.2, 2.4 μg) of monoclonal IgG 4C11 (lanes 3–6) or of non-immune rat IgG (lanes 7–10).