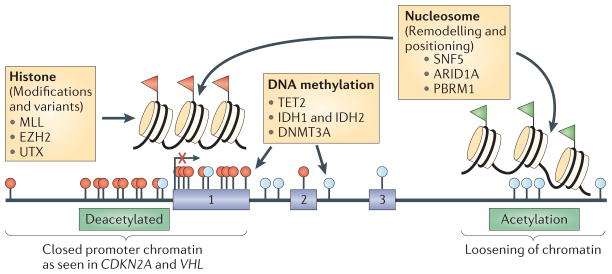
Figure 2. The cancer epigenome and relevant gene mutations.
The cancer epigenome is characterized by simultaneous global losses in DNA methylation (indicated by pale blue circles) with hundreds of genes that have abnormal gains of DNA methylation (indicated by red circles) and repressive histone modifications (indicated by red flags) in promoter region CpG islands. The hypomethylated regions have an abnormally open nucleosome configuration and abnormally acetylated histone lysines (indicated by green flags). Conversely, abnormal DNA hypermethylation in promoter CpG islands is associated with nucleosomes positioned over the transcription start sites of the associated silenced genes (indicated by an arrow with a red X). Recent whole-exon sequencing of human cancers has shown a high proportion of mutations in genes in leukaemias, lymphomas, and ovarian, renal and pancreatic cancers, and rhabdomyosarcoma109–111,154–156 (indicated in yellow boxes), which are depicted as helping to mediate either abnormal DNA methylation, histone modifications and/or nucleosome remodelling100,107,108,118,155,157–165. ARID1A, AT-rich interactive domain-containing protein 1A; DNMT3A, DNA methyltransferase 3A; EZH2, ehancer of zeste 2; IDH1, isocitrate dehydrogenase 1; MLL, mixed lineage leukaemia; PBRM1, protein polybromo 1; SNF5, SWI/SNF-related, matrix associated, actin-dependent regulator of chromatin, subfamily B, member 1; VHL, Von Hippel–Lindau.