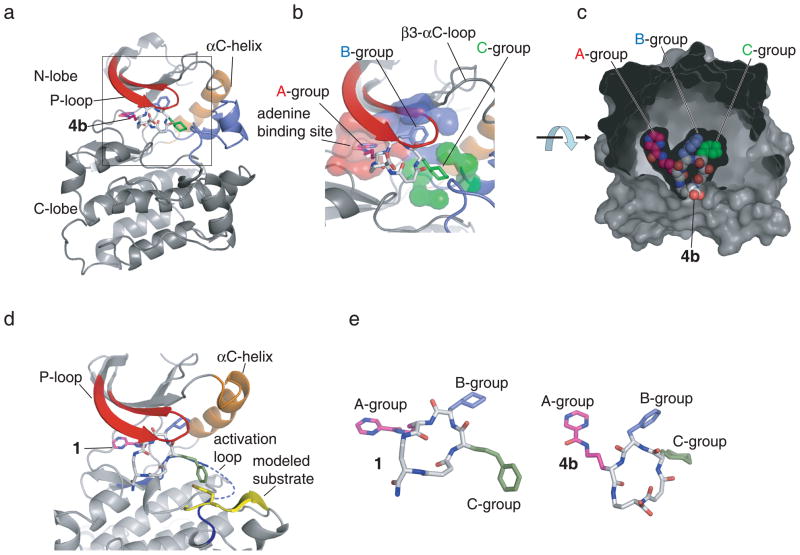
Figure 4.
The three-dimensional structure of Src kinase domain bound to macrocyclic inhibitors. (a) 4b binds to the active site of the kinase underneath the phosphate-binding P-loop (shown in red). The kinase adopts the Src/CDK-like inactive conformation characterized by the outward orientation of helix αC (orange) and the disruption of the peptide-binding patch by the activation loop (blue). (b) The three building block positions of the macrocycle compounds occupy three distinct binding pockets: the A-building block binds to the adenine binding pocket (red), the B-building block binds to a hydrophobic pocket underneath the β3 -αC loop, and the C-building block occupies a binding pocket (green) facing the Asp-Phe-Gly motif of the activation loop. (c) 4b binding pocket in detail. The surface created by the protein and ordered water molecules is shown in grey. The carboxylate group, from which DNA was attached during in vitro selection of the DNA-templated library and from which the fluorescein group was attached during binding affinity measurements, is exposed to solvent. (d) Superposition of the experimental X-ray crystal structure of Src•1 with the structure of the substrate peptide (yellow) from the complex with IRK (pdb-entry: 1IR3).32 (e) Comparison of the structures of 1 and 4b when complexed with Src kinase domain. The macrocycle structures are shown from a perspective that fixes the kinase domains (not shown) in the same orientation.