Figure 4.
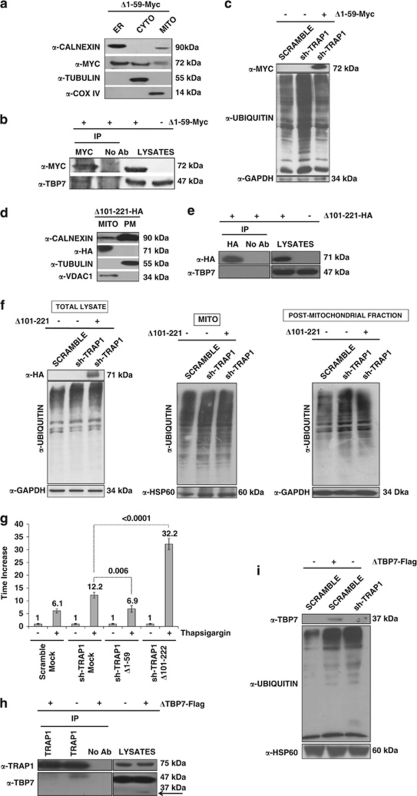
The TRAP1/TBP7 interaction in the ER is required for control of protein ubiquitination and ER stress. (a and d) Sub-cellular localization of Δ1–59-Myc/Δ101–221-HA mutants. HCT116 cells were transfected with the Δ1–59-Myc (a) or Δ101–221-HA (d) TRAP1 mutants; sub-fractionated into MITO, CYTO and microsomal (ER) fractions (a), or MITO and PM (cytosol+microsomes) fractions (d), as described under Materials and Methods; separated by SDS-PAGE; and immunoblotted using the indicated antibodies to verify the expression of mutants and the purity of fractions. For details on procedures for generation of the mutants see Materials and Methods. (b, e) Interaction between Δ1–59-Myc/Δ101–221-HA mutants and TBP7. HCT116 cells were transfected with Δ1–59-Myc (b) or Δ101–221-HA (e) TRAP1 mutants, harvested and immunoprecipitated using anti-Myc or anti-HA antibodies as described under Materials and Methods. Immunoprecipitates were separated by SDS-PAGE and immunoblotted using the indicated antibodies. No Ab, total cellular extracts incubated with A/G plus agarose beads without antibody; IP, immunoprecipitation with the corresponding antibodies. Three independent experiments were performed, with similar results. (c) Ubiquitination levels upon transfection of the Δ1–59-Myc TRAP1 deletion mutant. Total lysates from HCT116 scrambled, sh-TRAP1 stable clones and sh-TRAP1 cells transfected with the Δ1–59-Myc TRAP1 mutant were subjected to immunoblot analysis using mouse monoclonal anti-Ub antibodies to detect total ubiquitination levels and with an anti-GAPDH antibody for normalization of cell lysates. Three independent experiments were performed, with similar results. (f) Ubiquitination levels upon transfection of the Δ101–221-HA TRAP1 deletion mutant. HCT116 scramble, sh-TRAP1 and sh-TRAP1 cells transfected with the Δ101–221-HA TRAP1 mutant were sub-fractionated in PM (microsomes+CYTO fraction) and MITO fractions as described under Materials and Methods. Total lysates from the same cells were used as controls (left panel). Protein lysates were subjected to immunoblot analysis using mouse monoclonal anti-Ub antibodies to detect total ubiquitination levels. The purity of fractions was verified using mouse monoclonal anti-GAPDH (left and right panels) and mouse-monoclonal anti-COX IV (middle panel) antibodies. Three independent experiments were performed, with similar results. (g) Real-time RT-PCR analysis of BiP/Grp78 mRNA expression in scrambled and sh-TRAP1 HCT116 cells exposed to 1 μM TG for 12 h (same as in Figure 2b) and in sh-TRAP1 HCT116 cells transfected with the Δ1–59-Myc or Δ101–221-HA TRAP1 mutant, as indicated, before treatment with TG. The P-values indicate the statistical significance between the BiP/Grp78 levels under the indicated conditions. (h) Interaction between TRAP1 and the ΔTBP7-Flag deletion mutant. HCT116 cells were transfected with the ΔTBP7-Flag deletion mutant, harvested and immunoprecipitated using anti-TRAP1 antibodies as described under Materials and Methods. Immunoprecipitates were separated by SDS-PAGE and immunoblotted using the indicated antibodies. No Ab, total cellular extracts incubated with A/G plus agarose beads without antibody; IP, immunoprecipitation with the corresponding antibodies. Three independent experiments were performed, with similar results. The arrow indicates the ΔTBP7-Flag mutant band. (i) Ubiquitination levels upon transfection of the ΔTBP7-Flag deletion mutant. Total lysates from HCT116 scrambled cells transfected with ΔTBP7-Flag mutant were subjected to immunoblot analysis using mouse monoclonal anti-Ub antibodies to detect total ubiquitination levels and with mouse monoclonal anti-HSP60 antibodies for normalization of cell lysates. Three independent experiments were performed, with similar results