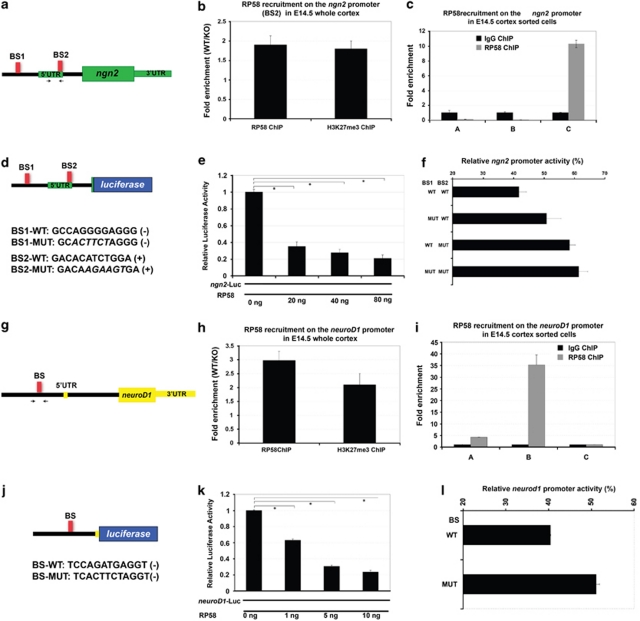
Figure 6.
RP58 directly regulates ngn2 and neuroD1 expression. (a) A schematic diagram showing the localization of two putative RP58-binding sites and the primers (arrrows) used in the ChIP–PCR in panels b and c. The ngn2 gene is indicated in green. The putative RP58-binding sites (BS1 and BS2) in the 5′ ngn2 genomic region are indicated by red boxes. (b) ChIP analyses of RP58 recruitment and presence of trimethylated histone H3K27 (H3K27me3) as a marker of repressed loci in control and RP58 mutant cortices using primers specific to the ngn2 genomic region encompassing the RP58-binding site BS2 at chr3:127336163+127336391 (see scheme in panel a). Note that the histograms show the fold enrichment of WT/KO. (c) Specific and preferential binding of RP58 to the ngn2 genomic region in the differentiated C cell populations. This experiment was performed on cells sorted from two independent litters, with primers encompassing the same binding site BS2, with similar results. Note that the background thresholds obtained with a ChIP experiment using an isotyped IgG are set at 1 (black bars). (d) Top: A schematic diagram showing the ngn2-luciferase construct (ngn2-luc plasmid: ngn2 regulatory region containing two RP58-binding sites) used for the data presented in panels e and f. Bottom: Sequences of the two RP58-binding sites, BS1 and BS2, in the ngn2 promoter region. Mutant versions of BS1 and BS2, and their sequence orientations are noted. (e) Transcriptional repression of an ngn2 regulatory region driving the expression of firefly luciferase by RP58. HEK293T cells were transfected with the ngn2-luc plasmid (see panel a). Ngn2-luc was transfected alone or with increasing doses of an RP58-expressing plasmid. Luciferase activity was assayed 48 h after transfection. Shown is a representative experiment of three independent biological replicates with similar results. The asterisks denote significant changes (P<0.001). (f) Mutation of RP58-binding sites impairs its repression of the ngn2 promoter. The ngn2-luc constructs containing no (WT), 1 or 2 mutant (MUT) RP58-binding sites were co-transfected into HEK293T cells together with an RP58-expressing plasmid. Luciferase activity was normalized as percentage of the same reporter activity in the absence of exogenous RP58. Shown is a representative experiment of three independent biological replicates with similar results. A statistically significant difference of activity was observed between BS1-MUT and BS2-MUT (P<0.01), as well as between the single mutant BS1-MUT and the double mutant BS1-MUT+BS2-MUT (P<0.01). (g) A schematic diagram showing the localization of the primers (arrows) used in the ChIP–PCR in panels h and i. The neuroD1 gene is indicated in yellow. The unique consensus RP58-binding site (BS) in the proximal 5′ neuroD1 genomic region is indicated by a red box. (h) ChIP analyses of RP58 recruitment and presence of trimethylated histone H3K27 (H3K27me3) as a marker of repressed loci in control and RP58 mutant cortices using primers specific to the neuroD1 genomic region encompassing the RP58-binding site. Note that the histograms show the fold enrichment of WT/KO. (i) Specific and preferential binding of RP58 to the neuroD1 genomic region in cells of the progenitor B cell population. Note that the background thresholds obtained with a ChIP experiment using an isotyped IgG are set at 1 (black bars). (j) Top: A schematic diagram showing the neuroD1-luciferase construct used for the data presented in panels k and l. Bottom: Sequence of the RP58-binding site BS in the neuroD1 promoter region. The mutant version of BS and sequence orientation is noted. (k) Transcriptional repression by RP58 of a neuroD1 regulatory region driving the expression of firefly luciferase. HEK293T cells were transfected with the neuroD1-luc plasmid (neuroD1 regulatory region containing one RP58-binding site driving a luciferase reporter gene). NeuroD1-luc was transfected alone or with increasing doses of an RP58-expressing plasmid. Shown is a representative experiment of three independent biological replicates with similar results. The asterisks denote significant changes (P<0.001). (l) Mutation of RP58-binding site impairs its repression of the neuroD1 promoter. The neuroD1-luc constructs containing the WT or mutant BS were co-transfected into HEK293T cells together with an RP58-expressing plasmid. Luciferase activity was normalized as percentage of the same reporter activity in the absence of exogenous RP58. Shown is a representative experiment of three independent biological replicates with similar results