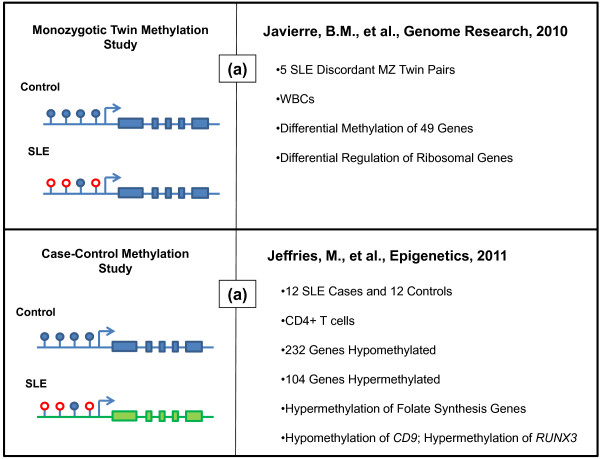
Figure 2.
Genome-wide methylation studies in lupus. (a) Differences in promoter DNA methylation were investigated in the context of identical genetic background (that is, discordant monozygotic (MZ) twins). Javierre and colleagues [61] performed a genome-wide methylation scan investigating epigenetic differences in five pairs of monozygotic twins discordant for lupus. The study investigated these differences in white blood cells (WBCs) of patients and their related controls. The study reported variable methylation of 49 of the 807 genes investigated. Pathway analysis of these genes reveals putative functions in immune response, cell activation, and cell proliferation. The study further identifies hypomethylation and overexpression of ribosomal genes, which may correspond to increased ribosomal-autoantibody formation. (b) Jeffries and colleagues [62] investigated the methylation status of over 27,000 individual CpG dinucleotides located in promoter regions of nearly 15,000 genes. Twelve lupus cases of varying disease activity and healthy control individuals were included in the study. The study reports hypomethylation of CpG dinucleotides in 232 genes and the hypermethylation of 104 genes. Pathway analysis reveals genes involved in folate biosynthesis, a pathway related to maintenance of DNA methylation. Further, modifications of individual genes observed to be dysregulated included the hypomethylation of CD9, encoding a known activator of T cell signaling, and hypermethylation of RUNX3, which encodes a transcription factor that mediates profileration signals in lymphocytes. In both of the diagrams on the left side of each panel, empty red circles represent unmethylated CpG dinucleotides, whereas solid blue circles represent methylated CpG sites. In the lower panel, the differences in color of the gene body indicate the presence of genetic heterogeneity.