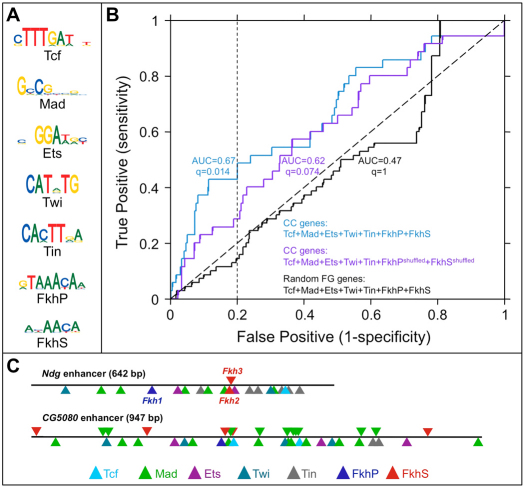
Fig. 6.
Enrichment of Fkh-binding sites in putative enhancers found within the noncoding regions of genes having heart expression. (A) Logo representations of the PWMs of the TFs used in the Lever analysis. (B) Receiver-operating characteristic (ROC) curves showing the discrimination of CC genes by the indicated AND combinations of Tcf, Mad, Ets, Twi, Tin and the two Fkh-binding motifs FkhP and FkhS. The area under the ROC curve (AUC) for each motif combination is shown. Note that Fkh motifs are over-represented in combination with motifs for all five known regulators of heart gene expression. Two other ROC curves are also shown to demonstrate that using shuffled instead of authentic Fkh motifs, or using a random gene set instead of the CC gene set, both increases the q-value and reduces the AUC. (C) Schematic of two of the three known CC enhancers that contain all seven of the TF-binding motifs used in the Lever analysis, including the Ndg enhancer used in this study (see also supplementary material Table S4).