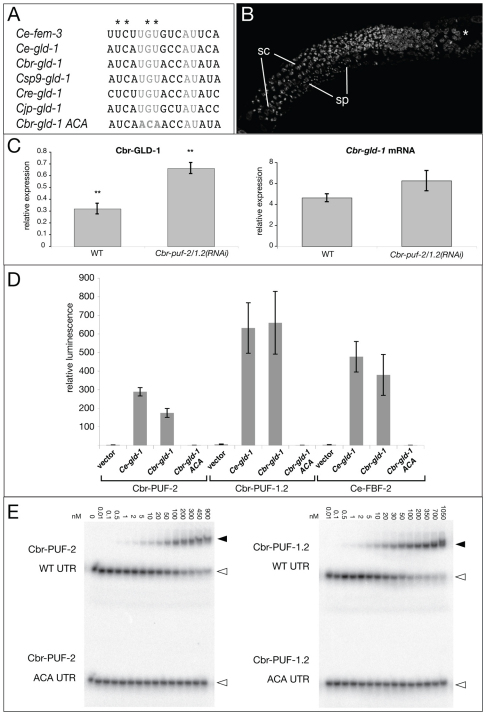
Fig. 4.
Cbr-gld-1 is a direct target of Cbr-PUF-2/1.2. (A) Alignment of FBF binding sites of C. elegans fem-3 and gld-1 with their orthologs from various Caenorhabditis species. Gray indicates the invariant core residues mutated in the ACA variant tested in D,E. Asterisks indicate fem-3(gf) (gain-of-function allele) point mutations. (B) Masculinization of germ line by Cbr-gld-1(RNAi);Cbr-puf-2/1.2(RNAi). Hoechst staining reveals spermatocytes (sc) at the gonad arm bend and highly condensed sperm (sp) nuclei at the proximal end of the gonad. Asterisk marks the distal tip of the gonad. (C) Cbr-GLD-1 level is significantly higher in Cbr-puf-2/1.2(RNAi) than in wild-type L4 worms, whereas the Cbr-gld-1 mRNA level is not. Error bars indicate s.e.; P=0.006 and P=0.168 for protein level and mRNA level, respectively (unpaired Student’s t-test). (D) Yeast three-hybrid interactions among C. elegans and C. briggsae PUF proteins and gld-1 mRNA variants. RNA plasmid pIIIa serves as the vector control. Error bars indicate s.e.m. (E) Cbr-PUF-2 and Cbr-PUF-1.2 bind to the putative Cbr-gld-1 FBE in vitro in a UGU-dependent manner. Black arrowheads indicate retarded complexes between labeled RNA oligomers and pure PUF proteins, and white arrowheads indicate free RNA oligomers.