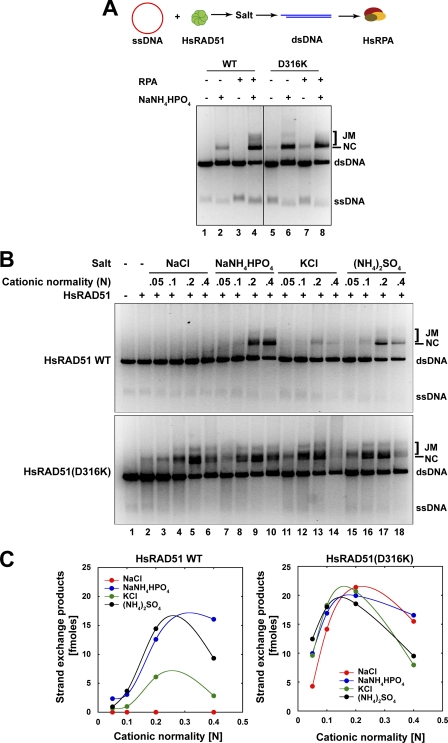
FIGURE 2.
HsRAD51(D316K) exhibits salt and RPA-independent strand exchange activity. A, analysis of salt and RPA requirement for strand exchange. Reaction schematic shown at top, HsRAD51 wild type or HsRAD51(D316K) (5 μm) and φX174 circular ssDNA (30 μm nt) were preincubated in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm of ATP at 37 °C for 5 min prior to the addition of 150 mm NaNH4HPO4 (if indicated) and linear φX174 dsDNA (15 μm bp). After 5 min, HsRPA (2 μm) was added (if indicated), and the incubation was continued for 3 h. Samples were deproteinized and analyzed on 0.9% agarose gel with 0.1 μg/ml ethidium bromide. Single-stranded DNA substrate (ssDNA), double-stranded DNA substrate (dsDNA), nicked circular product (NC), and joint molecule product (JM) are labeled on the right. B, strand exchange activity of HsRAD51 and HsRAD51(D316K) as a function of cation normality. HsRAD51 (5 μm) and φX174 circular ssDNA (30 μm nt) were preincubated with 2.5 mm ATP and 1 mm MgCl2 at 37 °C for 5 min before addition of indicated amounts of salt and linear φX174 dsDNA (15 μm bp). After another 5 min of incubation HsRPA (2 μm) was added, and the incubation was continued. After 3 h, samples were deproteinized and analyzed on 0.9% agarose gel with 0.1 μg/ml ethidium bromide. ssDNA, dsDNA, nicked circular product (NC), and joint molecule product (JM) are labeled on the right. C, quantification of the nicked circular (NC) + joint molecules (JM) products in B.