Background: UDP-galactopyranose mutase (UGM) catalyzes a step in galactofuranose biosynthesis in pathogens and is a promising drug design target.
Results: The first crystal structures and SAXS analysis of UGM from the pathogenic fungus Aspergillus fumigatus are reported.
Conclusion: The unique quaternary structure enables profound conformational changes to occur upon substrate binding. The structures support the covalent mechanism.
Significance: The structures should aid inhibitor design.
Keywords: Carbohydrate Biosynthesis, Flavoproteins, Protein Structure, X-ray Crystallography, X-ray Scattering
Abstract
UDP-galactopyranose mutase (UGM) is a flavoenzyme that catalyzes the conversion of UDP-galactopyranose to UDP-galactofuranose, which is a central reaction in galactofuranose biosynthesis. Galactofuranose has never been found in humans but is an essential building block of the cell wall and extracellular matrix of many bacteria, fungi, and protozoa. The importance of UGM for the viability of many pathogens and its absence in humans make UGM a potential drug target. Here we report the first crystal structures and small-angle x-ray scattering data for UGM from the fungus Aspergillus fumigatus, the causative agent of aspergillosis. The structures reveal that Aspergillus UGM has several extra secondary and tertiary structural elements that are not found in bacterial UGMs yet are important for substrate recognition and oligomerization. Small-angle x-ray scattering data show that Aspergillus UGM forms a tetramer in solution, which is unprecedented for UGMs. The binding of UDP or the substrate induces profound conformational changes in the enzyme. Two loops on opposite sides of the active site move toward each other by over 10 Å to cover the substrate and create a closed active site. The degree of substrate-induced conformational change exceeds that of bacterial UGMs and is a direct consequence of the unique quaternary structure of Aspergillus UGM. Galactopyranose binds at the re face of the FAD isoalloxazine with the anomeric carbon atom poised for nucleophilic attack by the FAD N5 atom. The structural data provide new insight into substrate recognition and the catalytic mechanism and thus will aid inhibitor design.
Introduction
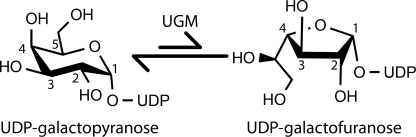
UDP-galactopyranose mutase (UGM)2 is a unique flavoenzyme that catalyzes the reversible conversion of UDP-galactopyranose (UDP-Galp) to UDP-galactofuranose (UDP-Galf) (Fig. 1). Unlike flavin-dependent oxidoreductases, the redox state of the flavin in UGM is unchanged upon product formation (1). Although it is known that the FAD must be reduced for maximal catalytic activity, the precise role that the flavin plays in catalysis remains controversial.
FIGURE 1.
Reaction catalyzed by UGM.
UGM is also an attractive target for drug design. The carbohydrate moiety of the product of the reaction, galactofuranose (Galf), is the five-membered ring form of the common monosaccharide galactose. Galf has never been found in mammals and higher plants but is an essential building block of the cell wall and extracellular matrix of many bacteria, fungi, and protozoa (2). The UGM reaction is central to Galf biosynthesis. Gene deletion studies have shown that UGM is essential for mycobacterial growth (3) and contributes to the virulence of the pathogenic fungus Aspergillus fumigatus (4) and the protozoan parasite Leishmania major (5). In summary, the importance of UGM for the viability of many pathogens and its absence in humans make UGM a potential drug target.
This study focuses on the UGM from A. fumigatus. Aspergillus spp. are ubiquitous fungi that cause diseases ranging from allergic reactions and lung infections to sepsis and death (6). Acquired by inhalation of airborne spores, Aspergillus infections (mainly A. fumigatus) can lead to invasive pulmonary aspergillosis, particularly in immunocompromised persons (6–8). Invasive pulmonary aspergillosis is also an emerging serious infection in patients with chronic obstructive pulmonary disease (9). The overall case fatality rate associated with invasive aspergillosis is 58% (10). Also, bronchial colonization by A. fumigatus in people with asthma or cystic fibrosis causes allergic bronchopulmonary aspergillosis, a hypersensitivity lung disease that can lead to airway destruction and widening and scarring of the lung, resulting in significant morbidity and mortality (11). The ubiquity of the fungus, the increasing occurrence of Aspergillus infections in humans, and the high mortality rate associated with invasive pulmonary aspergillosis demonstrate the need for new anti-Aspergillus drugs.
Inhibition of A. fumigatus UGM (AfUGM) is a potential strategy for the development of new drugs to treat patients with aspergillosis. In Aspergillus, Galf is present in the galactomannan fraction of the cell wall, N- and O-glycans of secreted glycoproteins, glycosylphosphatidylinositol-anchored lipophosphogalactomannan, and sphingolipids (12). It is estimated that Galf constitutes 5% of the dry weight of A. fumigatus (12). Deletion of the UGM gene in A. fumigatus results in a thinner cell wall and increased susceptibility to drugs (4). Furthermore, the mutated strain exhibits attenuated virulence in a low-dose mouse model of invasive aspergillosis (4). In another study, deletion of the UGM gene resulted in a reduced growth phenotype, but virulence as measured in a higher dose mouse model was unimpaired (13). The different outcomes of the two deletion studies may reflect the different doses used, with the low-dose model generally considered to mimic more closely the in vivo situation (2). Because of the importance of UGM for the fitness of A. fumigatus, UGM is a promising target for the development of new drugs for adjunctive treatment for aspergillosis (2, 4).
Here we report four crystal structures of AfUGM along with an analysis of the oligomeric state and quaternary structure in solution using small-angle x-ray scattering (SAXS). The data reveal interesting differences from bacterial UGMs, including additional secondary structure elements, unique mode of oligomerization, and profound conformational changes induced by substrate binding. The structures should aid inhibitor design efforts.
EXPERIMENTAL PROCEDURES
Crystallization
Structure determination was problematic because of translational pseudosymmetry in crystals of native and mutant AfUGMs, as described in detail in the supplemental data. This difficulty was circumvented by using the double mutant enzyme K344A/K345A for structure determination. The kinetic constants of K344A/K345A are virtually identical to those of the native enzyme (supplemental Table S1), and the structures show that residues 344 and 345 are on the surface of the enzyme, far from the active site and oligomerization interfaces.
AfUGM mutant K344A/K345A (referred to hereafter as AfUGM) was expressed and purified as described for the native enzyme (14). Se-Met AfUGM was produced using the metabolic inhibition method (15).
Crystals were grown in sitting drops at room temperature using drops formed by mixing 1.5 μl of the protein stock solution with an equal volume of the reservoir. The protein stock solution consisted of 8 mg/ml protein in 25 mm HEPES, 125 mm NaCl, 5 mm l-cysteine, and 0.5 mm Tris(hydroxypropyl)phosphine at pH 7.5. The optimal reservoir contained 1.5 m ammonium sulfate and 0.1 m sodium acetate, pH 4.5. The crystals appeared as large, yellow hexagonal bipyramids. The crystals were cryoprotected by exchanging the mother liquor with 1.8 m ammonium sulfate, 0.1 m sodium acetate, pH 4.5, and 25% glycerol. The space group is P6522 with unit cell dimensions of a = 218 Å and c = 320 Å. Although a conventional estimate of the solvent content using the method of Matthews (16) yields the expectation of eight molecules in the asymmetric unit, there are, in fact, only four molecules present. The solvent content and VM are thus 75% and 4.9 Å3/Da.
Crystal Soaking
Crystals of reduced AfUGM with and without bound active site ligands were prepared by soaking the aforementioned crystals. Crystals of ligand-free reduced AfUGM were prepared by soaking crystals in the cryobuffer supplemented with 80 mm dithionite. At this concentration, the enzyme is fully reduced in solution (14). Indeed, the yellow color of the crystals was bleached upon soaking, indicating that the FAD is reduced. When the transformation was complete (2–3 min), the crystals were plunged into liquid nitrogen to trap the reduced state. Crystals of the reduced enzyme complexed with UDP were prepared by soaking with the cryobuffer supplemented with 80 mm dithionite and 40 mm UDP. The soaking time was ∼30 min. Crystals of the reduced enzyme complexed with UDP-Galp were prepared similarly using 80 mm dithionite and 100 mm UDP-Galp. As with the ligand-free enzyme, the crystals of the ligand complexes were colorless when frozen.
X-ray Diffraction Data Collection, Phasing, and Refinement
Crystals were analyzed at beamlines 19ID and 24-ID-C of the Advanced Photon Source and beamline 8.2.2 of the Advanced Light Source. The 19ID data sets were processed with HKL3000 (17). The other data sets were integrated with XDS and scaled with SCALA (18) via CCP4i (19).
The first structure of AfUGM was determined using single-wavelength anomalous diffraction phasing based on data collected at 19ID from Se-Met AfUGM crystals (Table 1). Several single-wavelength anomalous diffraction data sets were collected at the wavelength corresponding to the experimentally measured peak of f″. The phasing potential of each data set was assessed with the HKL2MAP interface (20) to the SHELXC/D/E programs (21, 22). A 2.65 Å resolution data set with good anomalous signal was identified, from which a 56-atom selenium constellation was deduced. This constellation was input to PHENIX AutoSol (23) for single-wavelength anomalous diffraction phasing, density modification, and automated building. After density modification, the figure of merit improved from 0.42 to 0.77 for all reflections to 2.65 Å resolution. The model from automated building consisted of 1956 residues, 1778 of which were assigned to sequence. This model was adjusted and extended manually in COOT (24), aided by the density-modified single-wavelength anomalous diffraction map, and subsequently refined against a 2.35 Å resolution native data set using PHENIX (25). Several additional rounds of building and refinement were performed. The resulting structure provided the starting coordinates for the refinements of the other structures reported here. Data collection and refinement statistics and Protein Data Bank (PDB) deposition codes are listed in Table 1.
TABLE 1.
X-ray diffraction data collection and refinement
Values for the outer resolution shell of data are given in parentheses. APS, Advanced Photon Source; ALS, Advanced Light Source.
| Se-Met | Sulfate complex | AfUGMr | AfUGMr-UDP | AfUGMr-UDP-Galp | |
|---|---|---|---|---|---|
| Beamline | APS 19ID | APS 19ID | ALS 8.2.2 | ALS 8.2.2 | APS 24-ID-C |
| Active site ligand | Sulfate | Sulfate | None | UDP | UDP-Galp |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 |
| Unit cell parameters (Å) | a = 218.1, c = 319.1 | a = 217.8, c = 319.7 | a = 217.9, c = 322.8 | a = 218.2, c = 320.8 | a = 219.1, c = 322.4 |
| Wavelength | 0.97915 | 0.97915 | 1.00000 | 1.00000 | 0.97949 |
| Resolution (Å) | 55.00-2.65 (2.74-2.65) | 50.0-2.35 (2.43-2.35) | 48.6-2.25 (2.37-2.25) | 48.6-2.25 (2.37-2.25) | 48.8-2.25 (2.37-2.25) |
| Observations | 1,833,668 | 900,659 | 1,560,709 | 1,036,765 | 1,670,151 |
| Unique reflections | 128,441 | 167,455 | 211,192 | 205,755 | 207,614 |
| Rmerge (I) | 0.087 (0.351) | 0.086 (0.568) | 0.097 (0.599) | 0.086 (0.676) | 0.097 (0.599) |
| Rmeas (I) | 0.104 (0.643) | 0.096 (0.753) | 0.104 (0.645) | ||
| Rpim (I) | 0.038 (0.231) | 0.041 (0.323) | 0.035 (0.232) | ||
| Mean I/σ | 32.3 (5.5) | 20.1 (3.0) | 16.2 (3.5) | 17.6 (2.8) | 13.9 (3.4) |
| Completeness (%) | 99.0 (99.6) | 90.5 (91.1) | 99.8 (100.0) | 97.9 (99.7) | 97.3 (97.6) |
| Multiplicity | 14.3 (12.7) | 5.4 (5.4) | 7.4 (7.4) | 5.0 (5.0) | 8.0 (7.0) |
| No. of protein chains | 4 | 4 | 4 | 4 | |
| No. of protein residues | 2018 | 2020 | 2019 | 2019 | |
| No. of protein atoms | 15,597 | 15,580 | 15,551 | 15,562 | |
| No. of FAD atoms | 212 | 212 | 212 | 212 | |
| No. of ligand atoms | 0 | 0 | 50 | 72 | |
| No. of water molecules | 516 | 813 | 657 | 659 | |
| Rcryst | 0.188 (0.249) | 0.190 (0.244) | 0.192 (0.260) | 0.185 (0.249) | |
| Rfreea | 0.218 (0.297) | 0.215 (0.281) | 0.218 (0.297) | 0.210 (0.294) | |
| r.m.s.d.b bond lengths, Åc | 0.007 | 0.007 | 0.007 | 0.007 | |
| r.m.s.d.b bond angles, ° | 1.06 | 1.04 | 1.07 | 1.08 | |
| Ramachandran plotd | |||||
| Favored (no. of residues) | 1985 | 1983 | 1979 | 1977 | |
| Allowed (no. of residues) | 25 | 29 | 32 | 34 | |
| Outliers (no. of residues) | 0 | 0 | 0 | 0 | |
| Average B-factor (Å2) | |||||
| Protein | 35 | 30 | 33 | 35 | |
| FAD | 32 | 25 | 30 | 31 | |
| Active site ligand | 56 | 26 | 39 | ||
| Water | 34 | 34 | 34 | 37 | |
| Coordinate error (Å)e | 0.33 | 0.32 | 0.32 | 0.32 | |
| PDB code | 3UTE | 3UTF | 3UTG | 3UTH |
Small-angle X-ray Scattering
SAXS experiments were performed at the SIBYLS beamline (12.3.1) of the Advanced Light Source (26). Prior to analysis, a sample of AfUGM at 9.4 mg/ml was dialyzed into a buffer of 20 mm HEPES, 45 mm NaCl, 0.5 mm Tris(hydroxypropyl)phosphine at pH 7.5. Scattering intensities (I) were measured at three nominal protein concentrations to ensure concentration-independent scattering. Exposures of 0.5, 1.0, and 5.0 s were used to check for radiation damage. The scattering curves collected from the protein sample were corrected for background scattering using intensity data collected from the dialysis buffer. A composite scattering curve was generated with PRIMUS (27) by scaling and merging the background-corrected high q region data from the 5.0-s exposure with the low q region data from the 0.5-s exposure. The pair distribution function was calculated with GNOM (28). GASBOR (29) was used to calculate shape reconstructions, and DAMAVER (30) was used to average and filter the resulting dummy atom models. The Situs module pdb2vol was used to convert the averaged, filtered models into volumetric maps (31). SUPCOMB was used to superimpose the crystallographic tetramer onto the dummy atom model (32).
Kinetics
The mutase activity was tested with UDP-Galf as the substrate following procedures previously described (14). The enzyme concentration was determined using the flavin extinction coefficient at 450 nm of 10.6 mm−1cm−1 (14).
Structure Analysis
SSM (33) was used for structure superposition. The PDBePISA server was used to analyze protein interfaces in crystal lattices (34). COOT and PyMOL (35) were used to analyze noncovalent interactions.
RESULTS
Overall Fold and Flavin Binding Site
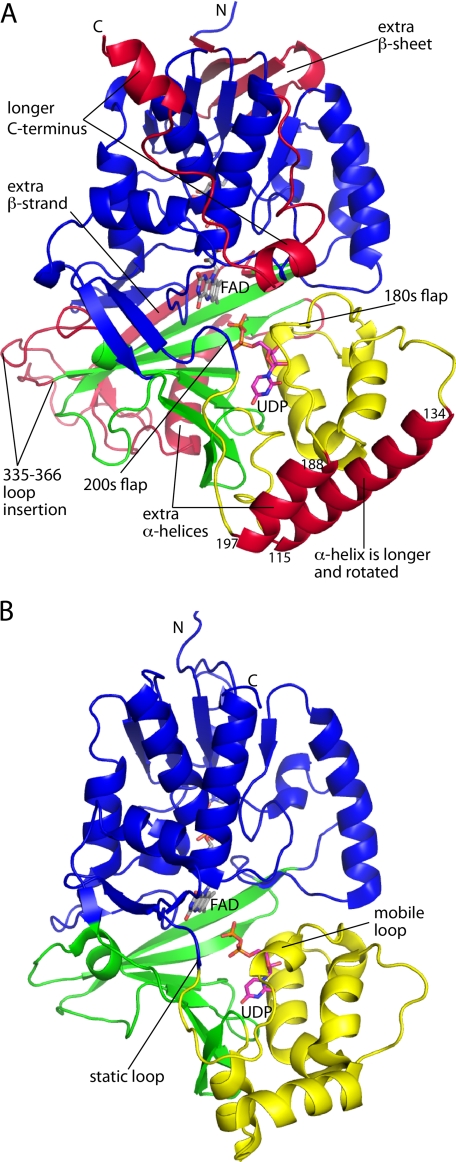
The AfUGM protomer has a mixed α/β-fold (Fig. 2) and exhibits the same three-domain architecture that was first identified in the structure of Escherichia coli UGM (36). Domain 1 is the largest of the three domains. It is a tripartite unit consisting of residues 3–90, 205–291, and 421–507 and includes a Rossmann fold core that binds the FAD. Domain 2 is a bundle of α-helices (residues 105–204). This domain mediates one of the major interfaces of the tetramer. Domain 3 (residues 91–104, 292–420) is situated between the other two domains and features a twisted, seven-stranded β-sheet. This domain participates in substrate binding. The FAD isoalloxazine binds in a crevice between domains 1 and 3.
FIGURE 2.
Overall fold of AfUGM. A, structure of the AfUGM protomer. Domains 1, 2, and 3 are colored blue, yellow, and green, respectively. Features that distinguish AfUGM from bacterial UGMs are colored red. B, protomer structure of a bacterial UGM (D. radiodurans UGM, PDB code 3HE3). This figure and others were prepared with PyMOL.
AfUGM is about 100 residues longer than bacterial UGMs, and the extra residues form several structural elements that appear to be important for catalysis and oligomerization (Fig. 2A, red sections). The first of these extra elements is a four-stranded β-sheet in domain 1 formed by residues 6–8 and 243–262. This β-sheet is located near the adenine of the FAD. Domain 2 has an extra helix (residues 188–197) not found in bacterial UGMs. This helix is situated between two mobile flaps that close and open in response to substrate binding and product release, respectively (Fig. 2A). Also, the second helix of the domain (residues 115–134) is about seven residues longer in AfUGM and rotated by about 90° from the corresponding helix in bacterial UGMs. The 115–134 and 188–197 helices form a four-helix bundle in one of the dimer interfaces of the UGM tetramer (see below). Domain 3 of AfUGM has two large inserts when compared with bacterial UGMs. The first is a long loop formed by residues 335–366. The second consists of residues 378–418, which fold into an α-helix followed by a β-strand. Because of the latter secondary structure element, the sheet of domain 3 has seven strands rather than six as in bacterial UGMs. Finally, AfUGM has ∼30 extra residues at the C terminus. These residues form a U-shaped substructure that packs against domain 1 and features two α-helices. This extra structural element is involved in oligomerization (see below).
Quaternary Structure
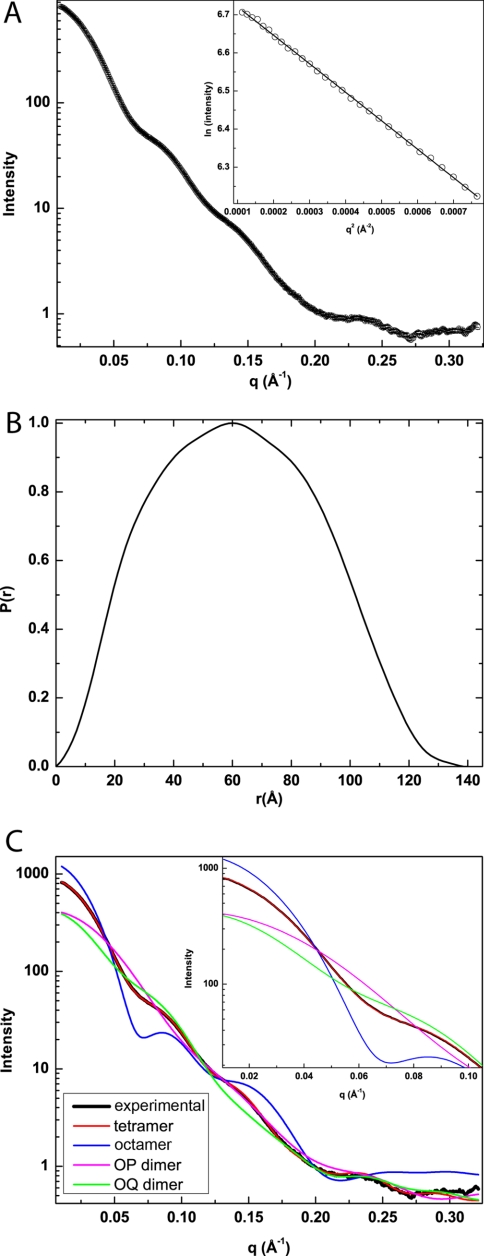
SAXS was used to obtain solution structural information for AfUGM. A composite scattering curve is shown in Fig. 3A. The associated Guinier plot exhibits good linearity (R2 = 0.9996) and suggests a radius of gyration of 47.3 Å (Fig. 3A, inset). Calculations of the pair distribution function yield a Rg of 47.6 Å and maximum particle dimension of 140 Å (Fig. 3B). For reference, the Rg of the crystallographic protomer is only 24 Å. These results suggest that AfUGM self-associates in solution.
FIGURE 3.
SAXS analysis. A, SAXS curve and Guinier plot (inset). The range of the Guinier plot corresponds to qRg = 0.503–1.31. B, pair distribution function. C, comparison of the experimental SAXS curve (black) with theoretical curves calculated from various oligomeric models of AfUGM. The inset highlights the fits in the low q region (q < 0.1 Å−1).
The SAXS data were further analyzed to determine the oligomeric state and quaternary structure. The experimental scattering profile was compared with theoretical profiles calculated from models of AfUGM oligomers. These models were obtained by analyzing the protein interfaces in the crystal lattice to identify potentially stable assemblies. The calculations revealed an octamer with Rg of 52 Å, a tetramer exhibiting point group 222 symmetry and having an Rg of 47 Å, and two symmetric dimers with Rg values of 34 Å (OP dimer) and 43 Å (OQ dimer). The Rg data suggest that AfUGM forms a tetramer. Indeed, only the profile calculated from the tetramer shows good agreement with the experimental curve (Fig. 3C). Note in particular that the agreement is exceptional for the low q region (q < 0.1, Fig. 3C, inset). These results are consistent with size exclusion chromatography data, which suggested that AfUGM forms a tetramer in solution (14). Finally, shape reconstruction calculations performed with GASBOR assuming a 222 tetramer yield an envelope that is in good agreement with the crystallographic tetramer (Fig. 4). It is concluded that the tetramer identified in the crystal lattice represents the solution structure of AfUGM.
FIGURE 4.
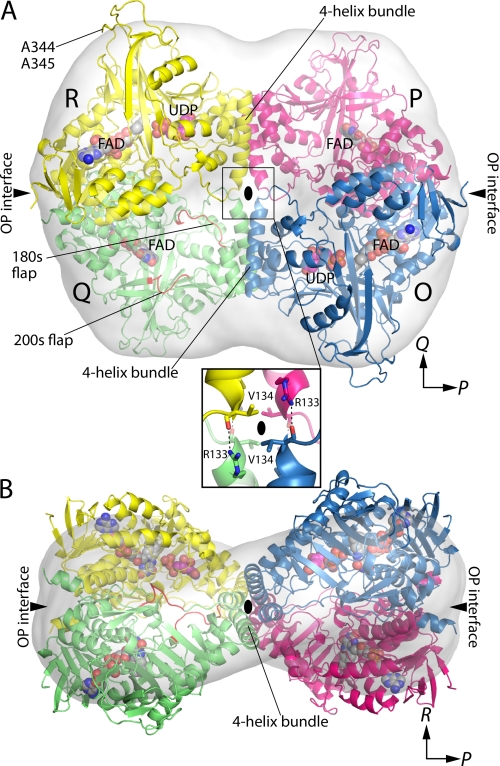
Quaternary structure of AfUGM. A, the tetramer is viewed down the R molecular two-fold axis. Each chain has a different color. The active site flaps of chain Q are colored red. The surface represents the SAXS shape reconstruction. Inset, intersubunit hydrogen bonds at the intersection of molecular two-fold axes. Chains related by the P-axis engage in hydrogen bonding. The oval represents the R two-fold molecular axis. B, the tetramer is viewed down the Q-axis.
The AfUGM tetramer is a dimer of dimers with point group 222 symmetry (Fig. 4). The symmetry of the tetramer can be described with three mutually orthogonal 2-fold axes, denoted P, Q, and R, that intersect at the center of the tetramer. The P-, Q-, and R-axes relate protomer O to protomers P, Q, and R, respectively. The axes define three potential dimer interfaces corresponding to protomer O contacting the other three protomers. Only the OP and OQ interfaces are significant.
The OP interface buries 5000 Å2 of area and is primarily hydrophilic, featuring 10 intersubunit hydrogen bonds. The interface is noncontiguous. The major part of the interface consists of the C-terminal residues 468–502 of two P-related chains interacting across the P-axis at the distal ends of the tetramer. Two such interfaces are located at opposite ends of the tetramer (Fig. 4). The minor part of the OP interface is located at the intersection of the two-fold axes and involves just the guanidinium of Arg-133 forming a hydrogen bond with the carbonyl of Val-134 of the P-related chain (Fig. 4A, boxed inset). Thus, there are four of these intersubunit hydrogen bonds located in the center of the tetramer.
The OQ interface is located in the middle of the tetramer and buries 4600 Å2 of contiguous surface area. Two helices (residues 115–134 and 188–197) from domain 2 of Q-related chains interact to form a four-helix bundle oriented parallel to the Q-axis (Fig. 4). Note that these two helices are unique to AfUGM (Fig. 2A). The tetramer has two of these bundles. The four-helix bundle has a hydrophobic interior featuring Val, Leu, and Ile side chains.
Active Site of Sulfate Complex
The crystals used for structure determination were grown in 1.5 m ammonium sulfate, and as crystallized, two sulfate ions are bound in the active site (supplemental Fig. S1A). The ions bind on the re side of the isoalloxazine. One of the ions binds next to the center ring of the isoalloxazine, which is the binding site for the Galp moiety of the substrate. His-63, an absolutely conserved residue in UGMs, is likewise located on the re face and interacts with both sulfate ions. The location of the conserved histidine at the re face is unprecedented and likely a crystallization artifact.
The isoalloxazine is planar, which is consistent with the FAD being oxidized. It is noted that the crystals are yellow, which is also indicative of the oxidized state.
Histidine Loop of Reduced AfUGM
The structure of the reduced enzyme (AfUGMr) was determined from a crystal that was soaked in 80 mm sodium dithionite. The crystal changed from yellow to colorless, which indicated that the FAD was reduced.
Soaking with dithionite causes significant changes in the active site, which occur in all four chains. Electron density features for the sulfate ions are absent, and the conformation of the histidine loop is dramatically different (supplemental Fig. S1B). In particular, His-63 has moved to the si face of the FAD, where it stacks in parallel with the middle ring of the isoalloxazine and forms a hydrogen bond with the FAD 2′-OH. Furthermore, the carbonyl of Gly-62 forms a hydrogen bond with the FAD N5. This conformational change in the protein is accompanied by a 3 Å shift in the FMN part of the FAD. This shift brings the N1 atom of the FAD within hydrogen-bonding distance of the backbone N-H group of Gln-458, which is consistent with the FAD adopting the anionic hydroquinone state.
The conformation of the histidine loop in AfUGMr is very similar to those of bacterial UGMs. In particular, in all other structures, the conserved histidine is located at the si face as in AfUGMr. Furthermore, in all other UGMs, the carbonyl of the residue preceding the conserved His accepts a hydrogen bond from the N5 of reduced FAD. The carbonyl-N5 interaction is important for stabilizing the reduced state of FAD, a necessity for activity. As mentioned, Gly-62 forms this critical hydrogen bond in AfUGMr. It is concluded that the dithionite-reduced AfUGM structure represents the active, substrate-free form of the enzyme.
FAD Conformation and Binding Site
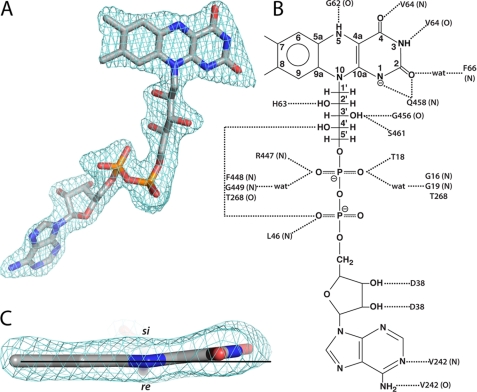
The FAD of AfUGMr binds primarily to domain 1 (Fig. 2A). The conformation of the FAD is nearly identical to those of bacterial UGMs, and the flavin-protein interactions are highly conserved (Fig. 5). In particular, there are several interactions with the protein backbone (Fig. 5B); these are conserved, as one would expect. Asp-38 forms two hydrogen bonds with the ribose. This interaction appears to be universal in UGM. Some of the side chain interactions with the FAD in AfUGM are found in other bacterial UGMs. For example, Ser-461 and Thr-18, which interact with the ribityl 3′-OH and pyrophosphate, respectively, are analogous to Thr-355 and Ser-14 of Klebsiella pneumoniae UGM (KpUGM).
FIGURE 5.
Electron density and interactions for FAD in AfUGMr. A, view of the FAD oriented with the re side of the isoalloxazine system facing the viewer. The cage represents a simulated annealing σA-weighted Fo − Fc omit map contoured at 3.0 σ. B, schematic diagram of protein-FAD interactions in AfUGMr. Backbone interactions are indicated by N or O in parentheses. C, edge-on view of the isoalloxazine. The horizontal line assists in seeing the 7° butterfly-like bend angle.
The FAD of AfUGMr exhibits a butterfly-like distortion of the isoalloxazine in which the pyrimidine ring bends 7° out of the plane such that the si face is concave (Fig. 5C). The direction of the bending is the same as that of Deinococcus radiodurans UGM (DrUGM) (37) and opposite to that of KpUGM (38). The angle is close to that of DrUGM and about half that of KpUGM (∼13°).
Structure of AfUGMr Complexed with UDP
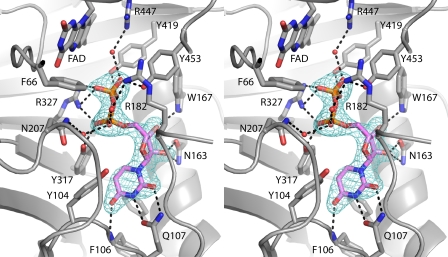
The structure of AfUGMr complexed with UDP was determined from a crystal that had been soaked in sodium dithionite and UDP. Electron density maps clearly indicated that UDP is bound in two of the four active sites of the tetramer (Fig. 6). The occupancy of UDP is estimated to be 0.9, which indicates essentially full occupancy and thus tight binding.
FIGURE 6.
Electron density map for UDP bound to AfUGMr (stereographic view). The cage represents a simulated annealing σA-weighted Fo − Fc omit map contoured at 3.0 σ.
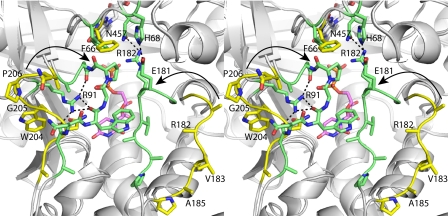
The binding of UDP induces profound conformational changes in the enzyme (Fig. 7). The largest of these changes involves residues 179–187 and 203–209. In the absence of UDP, these loops reside on the periphery of the enzyme, and thus, the active site is open. Upon binding, the loops move toward each other like the flaps of a box top to create a closed active site. The conformational changes are substantial. For example, the middle residues of the two loops, Val-183 and Pro-206, move by 11 and 13 Å, respectively, upon ligand binding. One consequence of the flaps folding inward is that the side chain of Phe-66 swings into the active site to avoid a clash with Pro-206 (Fig. 7). As shown below, Phe-66 contacts the Galp moiety of the substrate.
FIGURE 7.
Comparison of open and closed forms of AfUGMr (stereographic view). AfUGMr is shown in yellow. The AfUGMr-UDP complex structure is shown in green. UDP is colored pink. Selected interactions in the closed state are indicated by dashed lines. The arrows denote the direction of flap closure.
The closing of the flaps is accompanied by smaller conformational changes in residues 104–107 and 146–161 that fine-tune the uridine binding pocket. Tyr-104 and Phe-158 move apart from each other by 3 Å to create space for the incoming uracil. Phe-106 and Gln-107 shift by 1.0 and 2.0 Å, respectively, to form three hydrogen bonds with the base (interactions are shown in Fig. 6). Finally, movements of ∼1.0 Å bring Asn-163, Trp-167, Tyr-317, Arg-327, and Tyr-453 into contact distance of the ribose and pyrophosphate (Fig. 6).
The dramatic closing of the flaps also results in the formation of new protein-protein interactions that stabilize the closed active site conformation (Fig. 7). In particular, Glu-181 and Arg-182 of the 180s flap interact with His-68 and Asn-457, respectively, in the closed state, whereas in the open state, the interacting partners are separated by 17–19 Å. Another interaction that stabilizes the closed state involves Arg-91, whose side chain moves to make four hydrogen bonds with the 200s flap.
Structure of AfUGMr Complexed with UDP-Galp
The structure of AfUGMr complexed with the substrate UDP-Galp was determined from a crystal that was soaked in sodium dithionite and UDP-Galp. As with the UDP complex, electron density maps indicated binding in two of the four active sites (Fig. 8A). The occupancy of UDP-Galp is estimated to be 0.8. Electron density is weak for the O5 atom of the Galp moiety, suggesting the possibility of conformational disorder in this part of the ring. We note that conformational disorder of the Galp moiety has been observed previously ((38) PDB code 3INT). As seen with UDP, the substrate induces closure of both flaps.
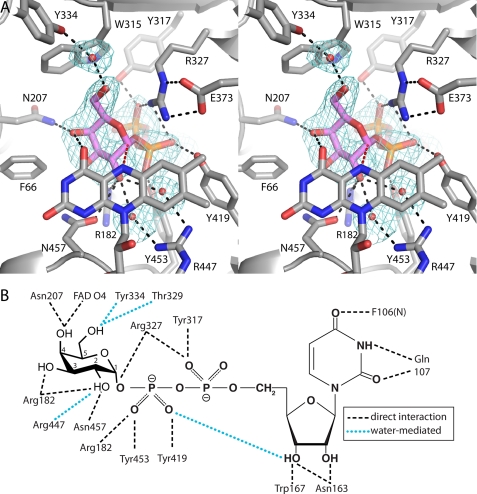
FIGURE 8.
Electron density map (stereographic view) and interaction diagram for UDP-Galp bound to AfUGMr. A, the cage represents a simulated annealing σA-weighted Fo − Fc omit map contoured at 2.5 σ. The red dashes indicate the 3.4 Å separation between the anomeric carbon atom of Galp and the FAD N5. B, schematic diagram of noncovalent electrostatic interactions.
The conformation and interactions of the UDP moiety of the substrate are essentially identical to those of the AfUGMr-UDP complex, so we will focus on the Galp moiety. Galp binds at the re face of the FAD isoalloxazine with the anomeric carbon atom (C1) poised 3.4 Å from the FAD N5 atom (Fig. 8A, red dashes). The hydroxyl groups of Galp make direct hydrogen bonds with Arg-182, Asn-207, and Asn-457 (Fig. 8B). Water-mediated interactions are formed with Thr-329, Tyr-334, and Arg-447. Also, the O4 hydroxyl forms a hydrogen bond with the FAD O4 carbonyl. In addition to these electrostatic interactions, nonpolar contacts are formed with Phe-66 and Trp-315. These steric interactions presumably help enforce specificity for Galp and orient the carbohydrate for catalysis.
Comparison of UDP-Galp Interactions in AfUGM and Bacterial UGMs
The structures of the UDP-Galp complexes of AfUGM, KpUGM, and DrUGM were compared to identify similarities and differences in substrate recognition (supplemental Fig. S2). In all three structures, the substrate adopts an extended conformation, the O4 hydroxyl of Galp forms a hydrogen bond to the FAD O4, and the anomeric carbon atom is near the FAD N5. Several conserved residues participate in substrate recognition, including Arg-182, Arg-327, Trp-167, Tyr-419, and Tyr-453 (AfUGM numbering).
Differences between the two classes of UGM are also evident (supplemental Fig. S2). In particular, the UMP moiety of AfUGM is displaced by 3–5 Å when compared with the bacterial enzymes. The uracil ring is shifted by ∼4 Å and rotated by almost 90°. This difference allows the uracil to form two hydrogen bonds with Gln-107, a residue without a counterpart in bacterial UGMs. Displacement of the UMP also reflects a different arrangement of Tyr residues that interact with the pyrophosphate (Tyr-317 in AfUGM, Tyr-185 in KpUGM, Tyr-209 in DrUGM). Finally, the O6 hydroxyl of Galp in AfUGM is rotated by 110° from that in the bacterial enzymes. This difference is due to the potential for steric clash with Trp-315.
DISCUSSION
The core structure of UGM appears to be conserved across the bacterial and eukaryotic domains of life despite low sequence identity (∼16%). The root mean square deviation between AfUGM and bacterial UGMs is 2.4–2.7 Å over 279–322 residues. Also, the majority of the FAD-protein interactions are conserved.
Curiously, protoporphyrinogen oxidase from Myxococcus xanthus (PPOX) (39), rather than another UGM, is the closest structural homolog of AfUGM in the PDB (supplemental Fig. S3). PPOX has FAD binding, membrane binding, and substrate binding domains, which correspond to UGM domains 1, 2, and 3, respectively. PPOX (PDB code 2IVD) and AfUGM superimpose with a root mean square deviation of 2.5 Å covering 370 residues, despite sharing only 15% sequence identity. Furthermore, PPOX has some of the extra structural features of AfUGM that are not found in bacterial UGMs, including the inserted β-sheet in domain 1 and the extra helix and strand of domain 3. The significance of the structural homology to PPOX is not obvious.
Neither oligomeric state nor quaternary structure is conserved in UGM. Dimers are common for bacterial UGMs, whereas AfUGM forms a tetramer in solution. UGM from E. coli was shown to be dimeric in solution based on dynamic light scattering (40), and the crystal structure revealed a semicircular dimer that likely represents the dimer in solution (36). This dimer is also found in the crystal lattices of KpUGM and Mycobacterium tuberculosis UGM (41). DrUGM appears to have a different quaternary structure, although this has not been confirmed with solution studies. Analysis of the interfaces in the primitive orthorhombic lattice of DrUGM suggests that the enzyme forms a decamer in solution (37). The decamer is a pentamer of dimers in which the propagated dimer is unlike the semicircular dimer of the other bacterial UGMs. The DrUGM dimer is formed by interactions involving domains 1 and 3. The constituent dimers of the AfUGM tetramer are unlike any of the dimers of bacterial UGMs.
Unique structural features of AfUGM preclude the assembly of the dimers formed by bacterial UGMs. For example, if the classic UGM semicircular dimer is constructed from AfUGM protomers, several clashes across the dimer interface are evident. These clashes involve the 200s flap and the extra helix of domain 2, both of which are unique to AfUGM. Likewise, a hypothetical dimer built like DrUGM exhibits clashes in the four-stranded sheet in domain 1 and the long inserted loop in domain 3. Thus, the protomer structure of AfUGM is incompatible with the quaternary structures of bacterial UGMs.
The observation of a tetramer raises the possibility of cooperativity between the subunits. Interestingly, previous studies have shown that 50% of the bound flavin remains reduced under normal laboratory conditions (absence of reducing agent), which suggests nonequivalence of the subunits (14). One possibility is that electrostatic repulsion between reduced flavins in the tetramer could contribute to this phenomenon. FADs related by the P-axis are 53 Å apart, whereas those related by the Q- and R-axes are separated by 75 and 79 Å, respectively. If electrostatic repulsion is important over the ∼50 Å scale, these distances suggest that protomers related by the R-axis, i.e. those along the diagonal of the tetramer, are equivalent. Interestingly, crystal soaking populated only the diagonal protomers. Future studies of cooperativity in AfUGM seem warranted.
The quaternary structure of AfUGM enables conformational changes that distinguish AfUGM from bacterial UGMs. AfUGM has two flaps that move substantially upon substrate binding and product release. Bacterial UGMs have just one mobile loop that enters the active site upon substrate binding (Fig. 2B, mobile loop) (37, 38). This loop is analogous to the 180s flap of AfUGM. The region of bacterial UGMs corresponding to the 200s flap also participates in substrate binding (Fig. 2B, static loop). In particular, Tyr-185 and Phe-186 of KpUGM (Tyr-209 and Phe-210 of DrUGM) contact the pyrophosphate and Galp moiety of UDP-Galp, respectively. However, these residues are static because the loop is in a protein-protein interface, and a neighboring protein molecule of the oligomer restricts the motion of the loop, effectively locking it in the closed conformation. In contrast, the 200s flap of AfUGM resides on the surface of the tetramer (Fig. 4A) and is able to adopt open and closed conformations. Thus, tetramerization of AfUGM allows an additional degree of conformation freedom.
It is notable that very few of the flap residues directly interact with the substrate. (Exceptions are Arg-182 and Asn-207.) This observation suggests that the purpose of flap closure is not only to assemble the constellation of residues needed for substrate recognition but also to create a protected environment for catalysis. The latter role is consistent with mechanisms in which UDP is displaced during the catalytic cycle (1) because closure of the flaps presumably prevents the severed UDP from migrating out of the active site.
Finally, the AfUGM structures provide additional insight into the catalytic mechanism of UGM. Two prevailing mechanisms have been proposed based on studies of the bacterial enzymes (1). One mechanism begins with the flavin functioning as a nucleophile that attacks the anomeric carbon of galactose to form a covalent intermediate and displace UDP (42) (supplemental Fig. S4). The other mechanism involves single electron transfer from the reduced flavin to an oxocarbenium sugar intermediate followed by the formation of a flavin-sugar adduct (43, 44). Which of these mechanisms, if either, applies to eukaryotic UGMs is unknown. However, the Galp moiety in the AfUGMr-UDP-Galp complex is poised for nucleophilic attack by the FAD N5 at the anomeric carbon atom (Fig. 8A). Also, the direction of the bend of the FAD isoalloxazine in AfUGM is consistent with formation of a covalent intermediate (41). For bacterial UGMs, crystal structures, reductive trapping experiments, and studies with modified flavins support the covalent mechanism (37, 38, 44). Thus, it seems likely that bacterial and eukaryotic UGMs share a common catalytic mechanism.
Supplementary Material
Acknowledgments
We thank Dr. Norma Duke, Dr. Greg Hura, and Dr. Jonathan Schuermann for help with x-ray data collection and processing. Part of this work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by Award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the United States Department of Energy, Office of Basic Energy Sciences, under Contract W-31-109-ENG-38. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the United States Department of Energy under Contract DE-AC02-05CH11231.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM094469 (to P. S. and J. J. T.).

This article contains supplemental crystallization methods, Table S1, and Figs. S1–S4.
The atomic coordinates and structure factors (codes 3UTE, 3UTF, 3UTG, and 3UTH) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- UGM
- UDP-galactopyranose mutase
- UDP-Galp
- UDP-galactopyranose
- UDP-Galf
- UDP-galactofuranose
- AfUGM
- UDP-galactopyranose mutase from A. fumigatus
- AfUGMr
- reduced form of UDP-galactopyranose mutase from A. fumigatus
- KpUGM
- UDP-galactopyranose mutase from K. pneumoniae
- DrUGM
- UDP-galactopyranose mutase from D. radiodurans
- SAXS
- small-angle X-ray scattering
- PDB
- Protein Data Bank
- Rg
- radius of gyration
- PPOX
- protoporphyrinogen oxidase from M. xanthus.
REFERENCES
- 1. Richards M. R., Lowary T. L. (2009) Chemistry and biology of galactofuranose-containing polysaccharides. Chembiochem 10, 1920–1938 [DOI] [PubMed] [Google Scholar]
- 2. Tefsen B., Ram A. F., van Die I., Routier F. H. (2011) Galactofuranose in eukaryotes: aspects of biosynthesis and functional impact. Glycobiology, in press [DOI] [PubMed] [Google Scholar]
- 3. Pan F., Jackson M., Ma Y., McNeil M. (2001) Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183, 3991–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmalhorst P. S., Krappmann S., Vervecken W., Rohde M., Müller M., Braus G. H., Contreras R., Braun A., Bakker H., Routier F. H. (2008) Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell. 7, 1268–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kleczka B., Lamerz A. C., van Zandbergen G., Wenzel A., Gerardy-Schahn R., Wiese M., Routier F. H. (2007) Targeted gene deletion of Leishmania major UDP-galactopyranose mutase leads to attenuated virulence. J. Biol. Chem. 282, 10498–10505 [DOI] [PubMed] [Google Scholar]
- 6. Kradin R. L., Mark E. J. (2008) The pathology of pulmonary disorders due to Aspergillus spp. Arch. Pathol. Lab. Med. 132, 606–614 [DOI] [PubMed] [Google Scholar]
- 7. Virnig C., Bush R. K. (2007) Allergic bronchopulmonary aspergillosis: a US perspective. Curr. Opin. Pulm. Med. 13, 67–71 [DOI] [PubMed] [Google Scholar]
- 8. Chong S., Lee K. S., Yi C. A., Chung M. J., Kim T. S., Han J. (2006) Pulmonary fungal infection: imaging findings in immunocompetent and immunocompromised patients. Eur. J. Radiol. 59, 371–383 [DOI] [PubMed] [Google Scholar]
- 9. Samarakoon P., Soubani A. O. (2008) Invasive pulmonary aspergillosis in patients with COPD: a report of five cases and systematic review of the literature. Chron. Respir. Dis. 5, 19–27 [DOI] [PubMed] [Google Scholar]
- 10. Lin S. J., Schranz J., Teutsch S. M. (2001) Aspergillosis case fatality rate: systematic review of the literature. Clin. Infect. Dis. 32, 358–366 [DOI] [PubMed] [Google Scholar]
- 11. Knutsen A. P., Slavin R. G. (2011) Allergic bronchopulmonary aspergillosis in asthma and cystic fibrosis. Clin. Dev. Immunol. 2011:843763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Latge J. P. (2009) Galactofuranose-containing molecules in Aspergillus fumigatus. Med. Mycol. 47, Suppl. 1, S104–S109 [DOI] [PubMed] [Google Scholar]
- 13. Lamarre C., Beau R., Balloy V., Fontaine T., Wong Sak Hoi J., Guadagnini S., Berkova N., Chignard M., Beauvais A., Latgé J. P. (2009) Galactofuranose attenuates cellular adhesion of Aspergillus fumigatus. Cell. Microbiol. 11, 1612–1623 [DOI] [PubMed] [Google Scholar]
- 14. Oppenheimer M., Poulin M. B., Lowary T. L., Helm R. F., Sobrado P. (2010) Characterization of recombinant UDP-galactopyranose mutase from Aspergillus fumigatus. Arch. Biochem. Biophys. 502, 31–38 [DOI] [PubMed] [Google Scholar]
- 15. Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 16. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 19. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 20. Pape T., Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 21. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 22. Sheldrick G. M. (2002) Macromolecular phasing with SHELXE. Z. Kristallogr. 217, 644–650 [Google Scholar]
- 23. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hura G. L., Menon A. L., Hammel M., Rambo R. P., Poole F. L., 2nd, Tsutakawa S. E., Jenney F. E., Jr., Classen S., Frankel K. A., Hopkins R. C., Yang S. J., Scott J. W., Dillard B. D., Adams M. W., Tainer J. A. (2009) Robust, high-throughput solution structural analyses by small-angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 28. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 29. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wriggers W. (2010) Using Situs for the integration of multiresolution structures. Biophys. Rev. 2, 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozin M. B., Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 33. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 36. Sanders D. A., Staines A. G., McMahon S. A., McNeil M. R., Whitfield C., Naismith J. H. (2001) UDP-galactopyranose mutase has a novel structure and mechanism. Nat. Struct. Biol. 8, 858–863 [DOI] [PubMed] [Google Scholar]
- 37. Partha S. K., van Straaten K. E., Sanders D. A. (2009) Structural basis of substrate binding to UDP-galactopyranose mutase: crystal structures in the reduced and oxidized state complexed with UDP-galactopyranose and UDP. J. Mol. Biol. 394, 864–877 [DOI] [PubMed] [Google Scholar]
- 38. Gruber T. D., Westler W. M., Kiessling L. L., Forest K. T. (2009) X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin. Biochemistry 48, 9171–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corradi H. R., Corrigall A. V., Boix E., Mohan C. G., Sturrock E. D., Meissner P. N., Acharya K. R. (2006) Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J. Biol. Chem. 281, 38625–38633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McMahon S. A., Leonard G. A., Buchanan L. V., Giraud M. F., Naismith J. H. (1999) Initiating a crystallographic study of UDP-galactopyranose mutase from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 55, 399–402 [DOI] [PubMed] [Google Scholar]
- 41. Beis K., Srikannathasan V., Liu H., Fullerton S. W., Bamford V. A., Sanders D. A., Whitfield C., McNeil M. R., Naismith J. H. (2005) Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidized state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state. J. Mol. Biol. 348, 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soltero-Higgin M., Carlson E. E., Gruber T. D., Kiessling L. L. (2004) A unique catalytic mechanism for UDP-galactopyranose mutase. Nat Struct. Mol. Biol. 11, 539–543 [DOI] [PubMed] [Google Scholar]
- 43. Fullerton S. W., Daff S., Sanders D. A., Ingledew W. J., Whitfield C., Chapman S. K., Naismith J. H. (2003) Potentiometric analysis of UDP-galactopyranose mutase: stabilization of the flavosemiquinone by substrate. Biochemistry 42, 2104–2109 [DOI] [PubMed] [Google Scholar]
- 44. Huang Z., Zhang Q., Liu H. W. (2003) Reconstitution of UDP-galactopyranose mutase with 1-deaza-FAD and 5-deaza-FAD: analysis and mechanistic implications. Bioorg. Chem. 31, 494–502 [DOI] [PubMed] [Google Scholar]
- 45. Engh R. A., Huber R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. Sect. A 47, 392–400 [Google Scholar]
- 46. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.