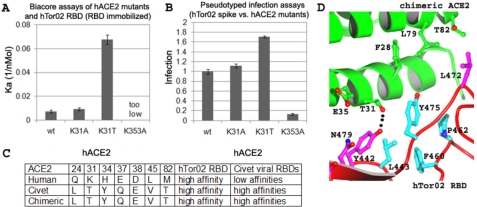
FIGURE 2.
Structures and functions of two virus-binding hot spots on hACE2. A, surface plasmon resonance Biacore analysis of the binding interactions between hTor02 RBD and wild-type or mutant hACE2. hTor02 RBD was immobilized, and wild-type or mutant ACE2 was flowed through. Each experiment was repeated six times at three different protein concentrations. The corresponding S.E. values are shown. B, pseudotyped viral infection assays of the interactions between hTor02 spike protein and wild-type or mutant hACE2. Retroviral murine leukemia viruses expressing β-galactosidase and pseudotyped with hTor02 spike protein were used to infect HEK293T cells expressing wild-type or mutant hACE2. Infection efficiency of pseudotyped viruses was measured by β-galactosidase assays and normalized against the infection efficiency in cells expressing wild-type hACE2. Each experiment was repeated six times. The corresponding S.E. values are shown. C, use of a human-civet chimeric ACE2 in crystallographic studies of RBD/cACE2 interactions. The chimeric ACE2 contains SARS-CoV-binding residues from cACE2 and other residues from hACE2. The chimeric ACE2 has the same receptor activities as cACE2 but the same crystallographic activities as hACE2 (27). SARS-CoV-binding residues that differ between hACE2 and cACE2 are shown. D, structure of the interface between hTor02 RBD and the chimeric ACE2.