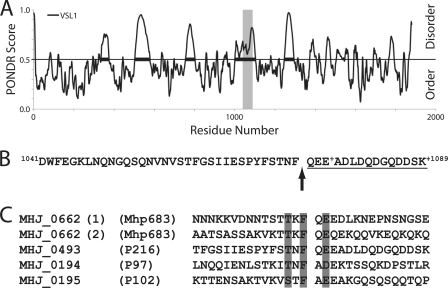
FIGURE 6.
Peptide motif TTKFQE found in Mhp683 is predictive of other cleavage sites within the P97/P102 family; identification of the Mhp493 cleavage site. A, PONDR analysis of Mhp493 (P216). Cleavage of Mhp493 into P120 and P85 was previously identified, but the precise cleavage site was unable to be accurately determined (15). Gray bar denotes region spanning amino acids 1041–1089 previously shown to house the putative cleavage site. B, amino acids 1041–1089 of the strain J homolog (MHJ_0493) of Mhp493. The N-terminal peptide identified by LC-MS/MS (supplemental Fig. 4) is underlined. Arrow denotes the site of cleavage that separates 85 kDa (P85) from the C terminus of MHJ_0493. + identifies a site of amino acid variability. The comparable sequence in Mhp493 from strain 232 is identical except an aspartic acid residue replaces a glutamic acid residue at position 1077 and an asparagine residue replaces a lysine residue at position 1089. C, comparison of identified P97/P102 family cleavage sites (14). Shaded regions denote highly similar or identical residues.