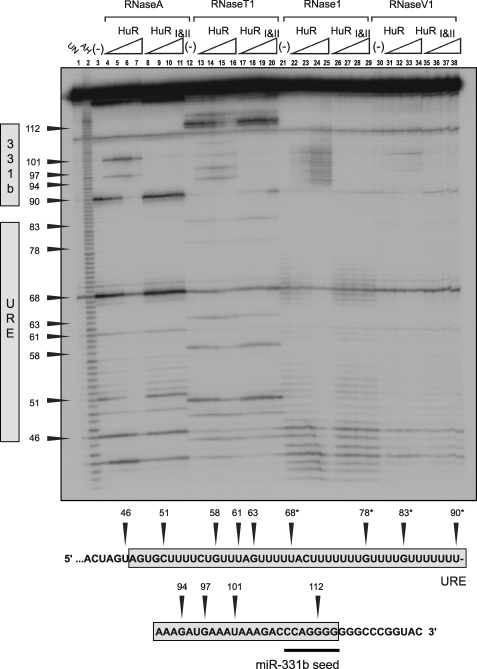
FIGURE 5.
RNase footprint analysis of HuR binding to ERBB-2 WT-URE/WT-331b RNA. Lane 1 contains “untreated” ERBB-2 WT-URE/WT-331b RNA (mock-digested) and is also labeled UN. Lane 2 contains ERBB-2 WT-URE/WT-331b RNA subjected to partial alkaline hydrolysis in order to generate a ladder corresponding to consecutive bases and is also labeled AH. Lanes 3–11 are partially digested with RNase A (which cleaves after U or C bases), and binding reactions contain the following: no protein (lane 3); 2 × 10−8 m HuR (lane 4); 5 × 10−8 m HuR (lane 5); 1 × 10−7 m HuR (lane 6); 2 × 10−7 m HuR (lane 7); 5 × 10−8 m HuRI&II (lane 8); 1 × 10−7 m HuRI&II (lane 9); 2 × 10−7 m HuRI&II (lane 10); or 5 × 10−7 m HuRI&II (lane 11). The presence or absence of protein is indicated as previously. RNase A digestion is also indicated by a label above lanes 3–11. Lanes 12–20 are RNase T1-digested (which cleaves after G bases); lanes 21–29 are RNase 1-digested (which cleaves preferentially after non-base-paired residues); and lanes 30–38 are RNase V1-digested (which cleaves preferentially after base-paired residues). Bands in the gel are assigned to the ERBB-2 WT-URE/WT-331b target RNA sequence below the gel by numbering. *, putative HuR binding site.