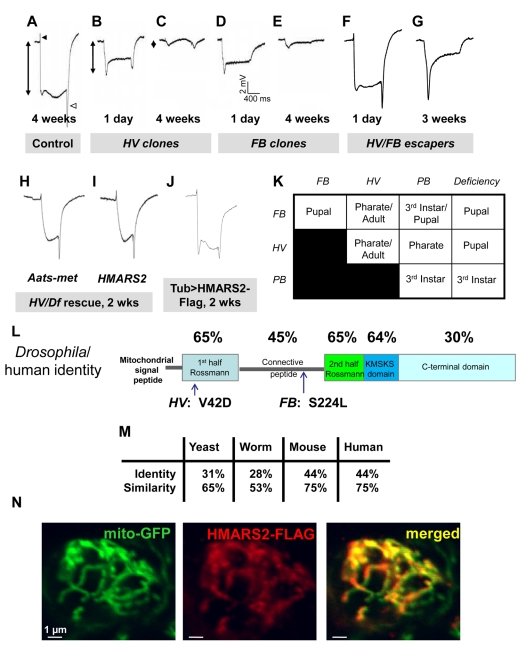
Figure 1. Identification/mapping of the Aats-met gene.
(A) ERG of the control (y w; FRT82B iso). The black and white arrowheads indicate the “on” and “off” transients, respectively. The double-pointed arrow indicates the amplitude. (B–C) ERGs of homozygous HV clone-containing flies at 1 d and 4 wk after eclosion. (D–E) ERGs of homozygous FB clone-containing flies at 1 d and 4 wk after eclosion. (F) ERG of a 1-d-old HV/FB escaper. (G) ERG of a 3-wk-old HV/FB escaper. (H) ERG of a 2-wk-old HV/Df fly rescued with actin-Gal4 and UAS-Aats-met. (I) ERG of a 2-wk-old HV/Df fly rescued with actin-Gal4 and UAS-HMARS2. (J) ERG of a 2-wk-old otherwise wild-type fly expressing HMARS2-FLAG driven by tub-Gal4. (K) Lethal stages of homozygous and transheretozygous allelic combinations reveal an allelic series: Df>PB>FB>HV. (L) The Aats-met protein's predicted domains are shown (drawn to scale), with position of mutations and percentage identity compared to human MARS2 shown. (M) The Drosophila Aats-met gene is homologous to the mitochondrial methionyl-tRNA synthetase genes of S. cerevisiae, C. elegans, M. musculus, and H. sapiens. (N) Colocalization of the Flag-tagged human MARS2 protein with Mito-GFP in the cell body of a neuron in the ventral nerve cord, driven by the D42-Gal4 driver, is shown.