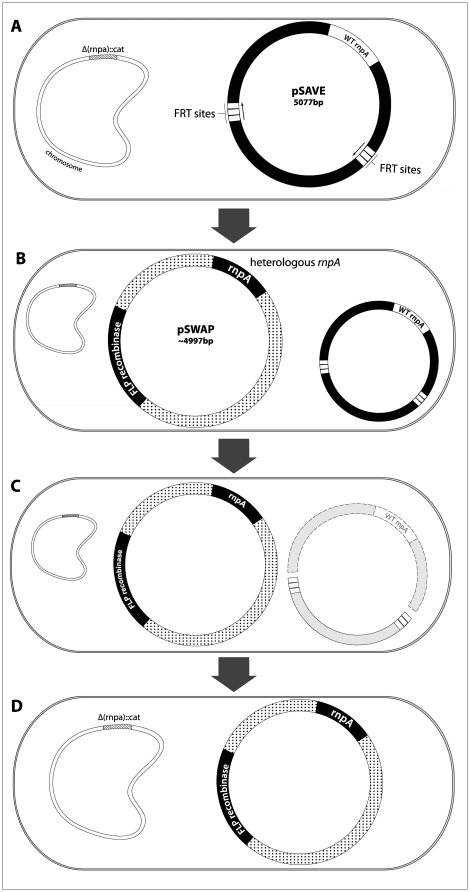
Figure 5. Complementation of E. coli with heterologous rnpA.
A) The chromosomal copy of rnpA has been deleted from lineage MTea1 and replaced by an antibiotic marker (cat) (crosshatched). Cells survive this deletion by relying on the pSAVE plasmid to express the heterologous RNase P protein. pSAVE has an inducible copy of the native E. coli rnpA under the control of a lacZ promoter; this region is flanked by two Flp-recombinase targets (FRT); B) Lineages relying solely on the heterologous rnpA for survival were constructed by inserting a second plasmid, pSWAP, into MTea1. Besides the heterologous rnpA, pSWAP contains the Flp recombinase gene; C) Since FRT sites in pSAVE are oriented in the same direction, the expression of the pSWAP FLP recombinase promotes the deletion of the pSAVE fragment harboring the E. coli rnpA and encompassed by the FRT sites (dashed). The fragments of pSAVE are subsequently lost; D) The resulting E. coli lineage survives with the heterologous rnpA present in pSWAP, provided that the heterolog is capable of rescuing RNase P functionality.