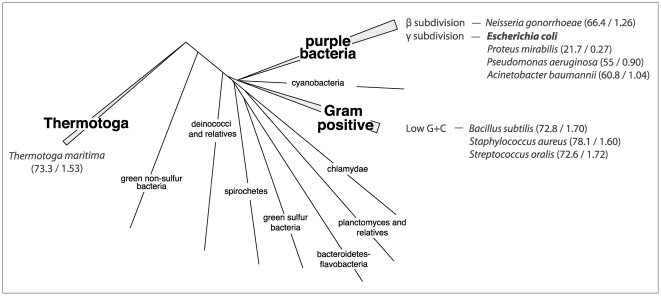
Figure 6. Phylogenetic range of rnpA heterologs investigated in this study.
The phylogenetic position and range of sources of heterologous rnpA cloned into pSWAP for complementation and fitness studies are shown on a consensus eubacterial phylogenetic tree. The sampled branches are shown in boldface, and the species listed in italics alongside. Escherichia coli (bold) was used as control. The two numbers in the parentheses quantify the pairwise divergence between the heterologous and the E. coli rnpA sequences: the first number is the unadjusted pairwise amino-acid divergence across the entire length of the sequence (expressed as a percentage), the second the corrected per-site divergence (number of amino-acid substitutions per amino-acid site using JTT substitution model as implemented in MEGA5.0 [51]). Eubacterial phylogenetic tree adapted from [47].