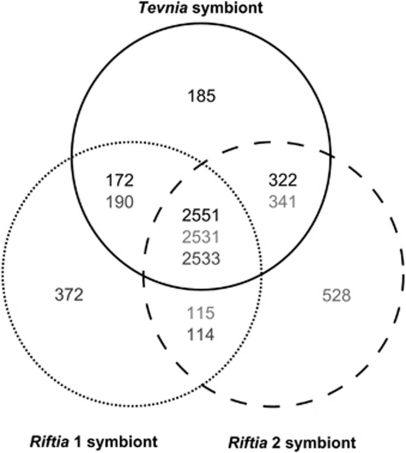
Figure 1.
Metagenome comparison. The Venn diagram depicts the number of genes shared (global alignment score >50%) or specific for the three symbionts. All bidirectional BLAST hits were globally screened by means of the Needleman–Wunsch algorithm. 2531–2551 open-reading frames (ORFs) are highly conserved among the three symbiont metagenomes and can be considered as orthologs (in the center). Variations in core genome size result from gene content redundancy in the metagenomes caused by paralogs and domain duplications. The number of ‘unique' genes is primarily attributed to the sequence fragmentation caused by repetitive elements and unavoidable host DNA contaminations. Black numbers, solid line: Tevnia symbiont; dark gray numbers, dotted line: Riftia 1 symbiont; light gray numbers, dashed line: Riftia 2 symbiont.