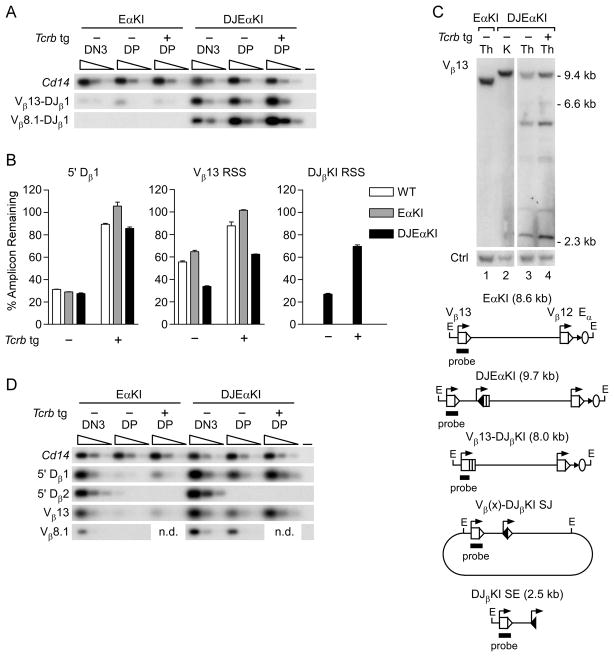
FIGURE 4.
Recombination of DJEαKI alleles. (A) Coding joint analysis. Genomic DNAs from sorted EαKI and DJEαKI DN3 and DP thymocytes without (−) or with (+) a Tcrb tg were serially three-fold diluted (wedges) and analyzed by PCR. Blots of PCRs using Vβ and Jβ1.1 primers were hybridized with a DJβKI-specific probe. Cd14 amplification was used to control for DNA loading. –, no DNA. Data are representative of two independent experiments. (B) RSS usage. Genomic DNAs from DJEαKI kidney and from sorted wild-type (WT), EαKI and DJEαKI DP thymocytes, without (−) or with (+) a Tcrb tg, were analyzed by quantitative real-time PCR. Percent amplicon remaining was calculated as [(experimental amplicon in thymus/experimental amplicon in kidney)/(B2m in thymus/B2m in kidney)] x 100. Data are the mean ± s.e.m. of 2–3 samples for each genotype. (C) Genomic Southern blot. Top, unfractionated thymus (Th) and kidney (K) genomic DNAs were digested with EcoRI and analyzed by Southern blot using a Vβ13 probe. DNA loading was assessed using a control (Ctrl) trypsinogen probe. Below, schematic of expected EcoRI fragments, including a diagram of predicted excision circles containing Vβ(x)-DJβKI signal joint (SJ) recombination products. (D) SE analysis. Thymocyte genomic DNA samples were linker-ligated, serially three-fold diluted (wedges) and analyzed by PCR. Cd14 amplification was used to control for DNA loading. –, no DNA. The data are representative of two independent experiments. n.d., not determined.