Abstract
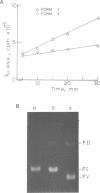
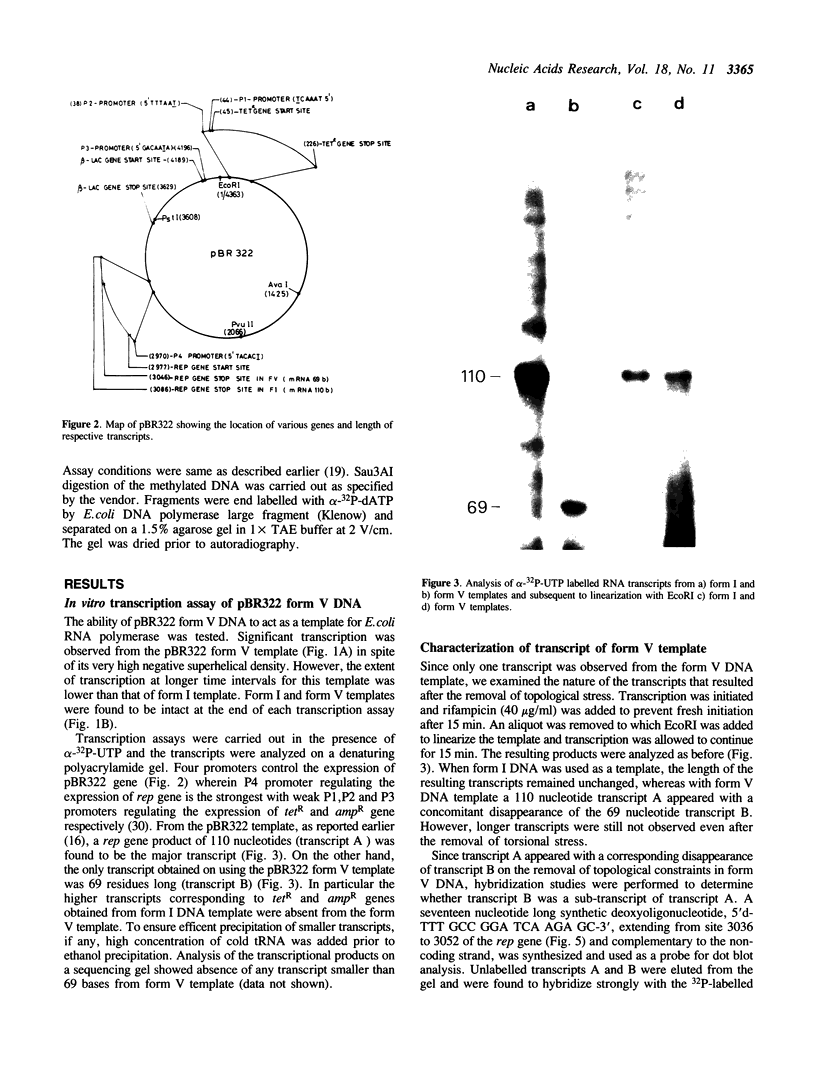
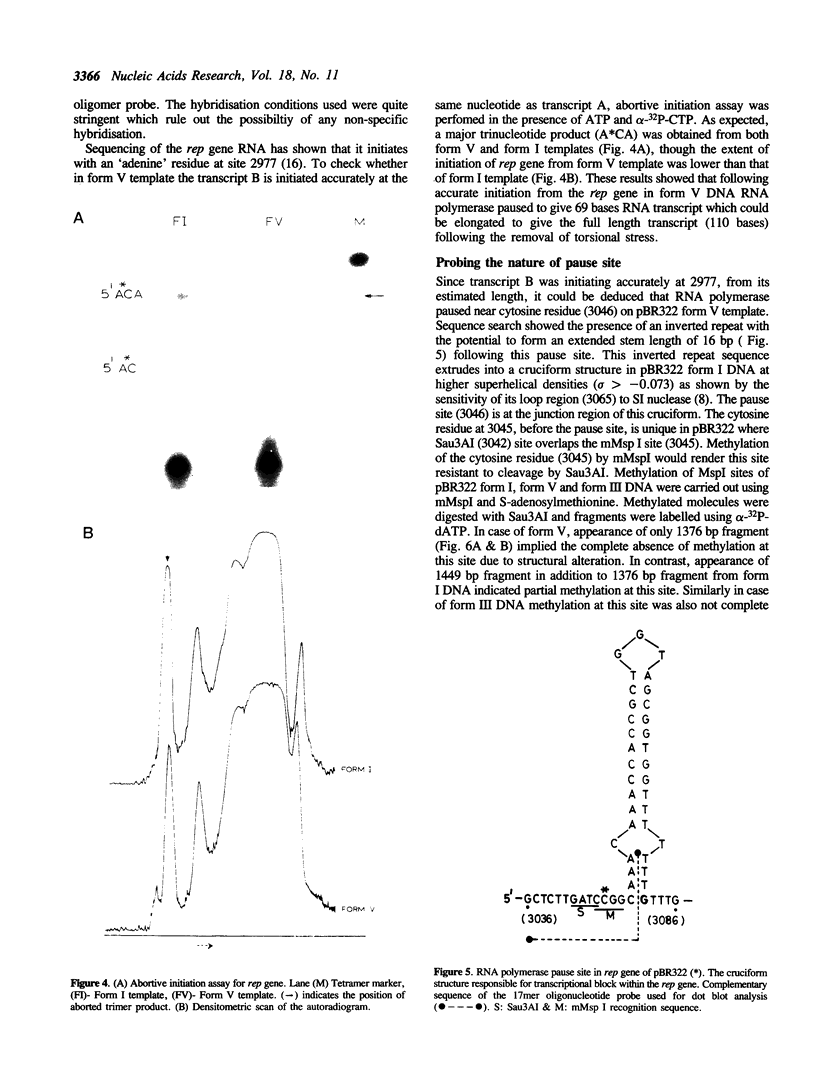
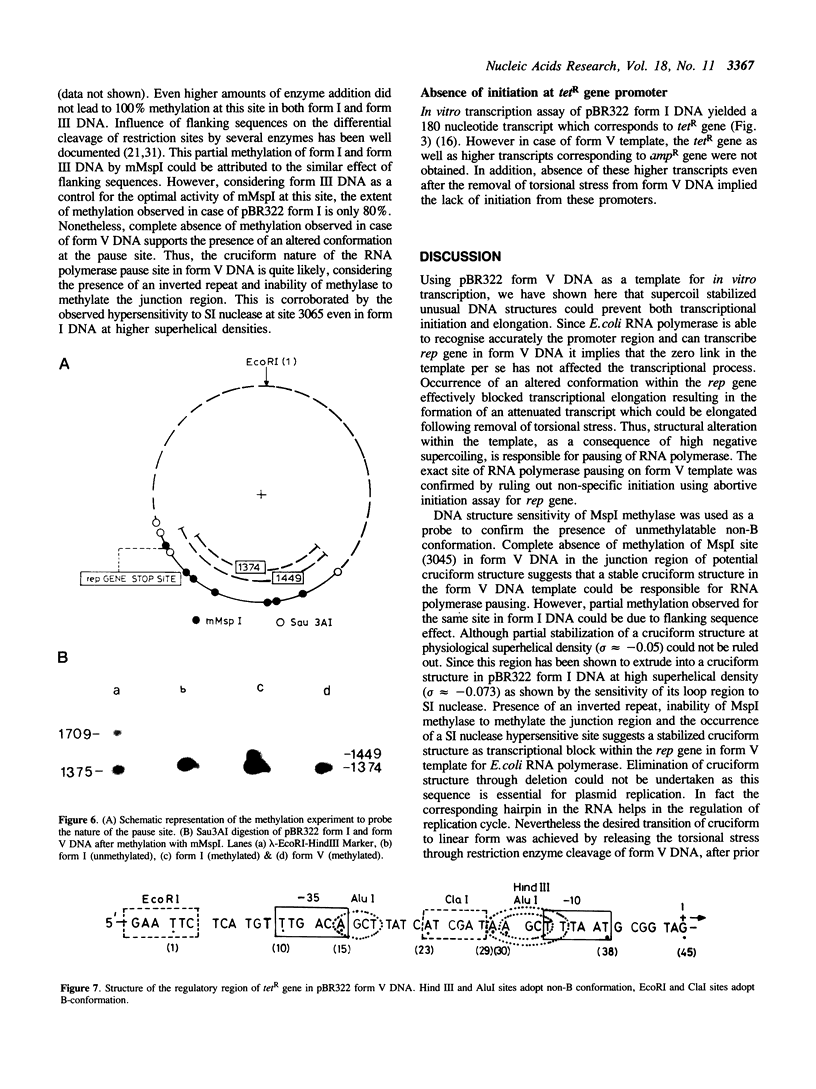
The transcriptional activity of pBR322 form V DNA template, a topologically unlinked, highly supercoiled molecule having unusual structures around or within coding regions was studied. Significant transcription was observed in vitro from this template despite high levels of supercoiling. An attenuated transcript, initiated accurately from the P4 promoter of rep gene, was observed which indicated pausing of E. coli RNA polymerase within the gene. This pausing could be removed by relieving the torsional stress implying that a supercoil induced structural alteration within the gene was acting as a transcriptional block. A stabilized unusual structure, most likely a cruciform, was found to be responsible for the elongation block. Absence of initiation from the tetR gene was correlated with the unusual structure present within its promoter region in form V DNA. These in vitro studies show that structural alterations within natural DNA could act as transcriptional blocks both at the level of initiation and elongation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E. Interaction of a protein from rat liver nuclei with cruciform DNA. EMBO J. 1988 Mar;7(3):843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. All three elements of the lac ps promoter mediate its transcriptional response to DNA supercoiling. J Mol Biol. 1987 May 5;195(1):89–97. doi: 10.1016/0022-2836(87)90329-9. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Brahmachari S. K., Mishra R. K., Bagga R., Ramesh N. DNA duplex with the potential to change handedness after every half a turn. Nucleic Acids Res. 1989 Sep 25;17(18):7273–7281. doi: 10.1093/nar/17.18.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S. K., Shouche Y. S., Cantor C. R., McClelland M. Sequences that adopt non-B-DNA conformation in form V DNA as probed by enzymic methylation. J Mol Biol. 1987 Jan 5;193(1):201–211. doi: 10.1016/0022-2836(87)90637-1. [DOI] [PubMed] [Google Scholar]
- Brahms J. G., Dargouge O., Brahms S., Ohara Y., Vagner V. Activation and inhibition of transcription by supercoiling. J Mol Biol. 1985 Feb 20;181(4):455–465. doi: 10.1016/0022-2836(85)90419-x. [DOI] [PubMed] [Google Scholar]
- Brahms S., Vergne J., Brahms J. G., Di Capua E., Bucher P., Koller T. Natural DNA sequences can form left-handed helices in low salt solution under conditions of topological constraint. J Mol Biol. 1982 Dec 5;162(2):473–493. doi: 10.1016/0022-2836(82)90539-3. [DOI] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. Structural junctions in DNA: the influence of flanking sequence on nuclease digestion specificities. Nucleic Acids Res. 1985 Jun 25;13(12):4445–4467. doi: 10.1093/nar/13.12.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. Gene transcription. Waves of DNA supercoiling. Nature. 1989 Jan 19;337(6204):206–206. doi: 10.1038/337206a0. [DOI] [PubMed] [Google Scholar]
- Gómez-Eichelmann M. C. Effect of nalidixic acid and novobiocin on pBR322 genetic expression in Escherichia coli minicells. J Bacteriol. 1981 Dec;148(3):745–752. doi: 10.1128/jb.148.3.745-752.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. An E. coli promoter that regulates transcription by DNA superhelix-induced cruciform extrusion. Science. 1988 Aug 5;241(4866):703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Worcel A. The positive transcription factor of the 5S RNA gene induces a 5S DNA-specific gyration in Xenopus oocyte extracts. Cell. 1985 Jul;41(3):945–953. doi: 10.1016/s0092-8674(85)80075-1. [DOI] [PubMed] [Google Scholar]
- Lafer E. M., Sousa R., Rosen B., Hsu A., Rich A. Isolation and characterization of Z-DNA binding proteins from wheat germ. Biochemistry. 1985 Sep 10;24(19):5070–5076. doi: 10.1021/bi00340a017. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra U., Barash F. DNA-dependent synthesis of RNA by Escherichia coli RNA polymerase: release and reinitiation of RNA chains from DNA templates. Proc Natl Acad Sci U S A. 1969 Oct;64(2):779–786. doi: 10.1073/pnas.64.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. R., Fitch W. M. Evolutionary selection for perfect hairpin structures in viral DNAs. Nature. 1982 Aug 5;298(5874):582–585. doi: 10.1038/298582a0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Thomae R., DiCapua E. Antibodies to Z-DNA interact with form V DNA. Nature. 1982 Dec 9;300(5892):545–546. doi: 10.1038/300545a0. [DOI] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Brahmachari S. K. Dynamic nature of B to Z transition: role of DNA supercoiling and Z-DNA binding protein. Indian J Biochem Biophys. 1988 Dec;25(6):542–547. [PubMed] [Google Scholar]
- Ramesh N., Shouche Y. S., Brahmachari S. K. Recognition of B and Z forms of DNA by Escherichia coli DNA polymerase I. J Mol Biol. 1986 Aug 20;190(4):635–638. doi: 10.1016/0022-2836(86)90248-2. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Santoro C., Costanzo F., Ciliberto G. Inhibition of eukaryotic tRNA transcription by potential Z-DNA sequences. EMBO J. 1984 Jul;3(7):1553–1559. doi: 10.1002/j.1460-2075.1984.tb02010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouche Y. S., Ramesh N., Brahmachari S. K. Probing of unusual DNA structures in topologically constrained form V DNA: use of restriction enzymes as structural probe. Nucleic Acids Res. 1990 Jan 25;18(2):267–275. doi: 10.1093/nar/18.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K. Effects of salts, temperature, and stem length on supercoil-induced formation of cruciforms. J Biol Chem. 1983 Jun 25;258(12):7661–7668. [PubMed] [Google Scholar]
- Smith G. R. DNA supercoiling: another level for regulating gene expression. Cell. 1981 Jun;24(3):599–600. doi: 10.1016/0092-8674(81)90085-4. [DOI] [PubMed] [Google Scholar]
- Stettler U. H., Weber H., Koller T., Weissmann C. Preparation and characterization of form V DNA, the duplex DNA resulting from association of complementary, circular single-stranded DNA. J Mol Biol. 1979 Jun 15;131(1):21–40. doi: 10.1016/0022-2836(79)90299-7. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Wright S., Bishop J. M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci U S A. 1989 Jan;86(2):505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]