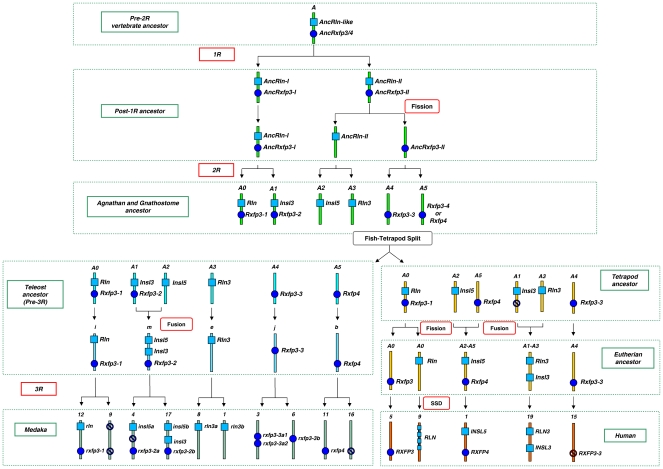
Figure 2. Reconstruction of the genetic events that led to the diversification of RXFP3-type receptors and RLN/INSL hormones in vertebrates based on the “fission” scenario of ancestral genome rearrangement.
The genomic origins of the hypothetical ancestral relaxin (AncRln-like) and Rxfp3/4 receptor (AncRxfp3/4) genes can be traced to a single chromosome in the vertebrate ancestor that had not yet been through 2R (Pre-2R vertebrate ancestor). The ancestral linkage group harboring AncRln-like and AncRxfp3/4-like sequentially underwent duplication, fission and another duplication yielding 5 distinct linkage groups (agnathan and gnathostome ancestor) harboring the ligand and receptor genes. Subsequently, tetrapods completely lost RXFP3-2 and often RXFP3-3 genes, but retained all of the post-2R ligand gene duplicates. Teleosts, on the other hand, retained both all of the ligand and receptor post-2R gene duplicates, suggesting that RXFP3-2 and RXFP3-3 acquired important functions in the pre-3R teleost ancestor. The duplicates of rxfp3-2 and rxfp3-3 were again retained in the post-3R teleost ancestor along with those of rln3 and insl5 (indicating their possible ligand-receptor relationships). Lastly, in vertebrates the RLN locus underwent multiple local duplications, resulting in the emergence of INSL4 in all eutherians, and INSL6 and RLN1 only in apes, whose RLN2 is orthologous to RLN of other eutherians. For simplicity, tetrapod and eutherian ancestor linkage groups are only shown to contain the fragments (e.g. A0, A2–A5) harboring the genes of interest; thus they should not be confused with actual chromosomes. Blue circles and squares represent receptor and their ligand genes respectively. Crossed circles represent pseudogenes (red, if they are verified in databases, blue if they are hypothetical). SSD: small-scale duplication. The first letter of ancestral gene names is capitalized.