Abstract
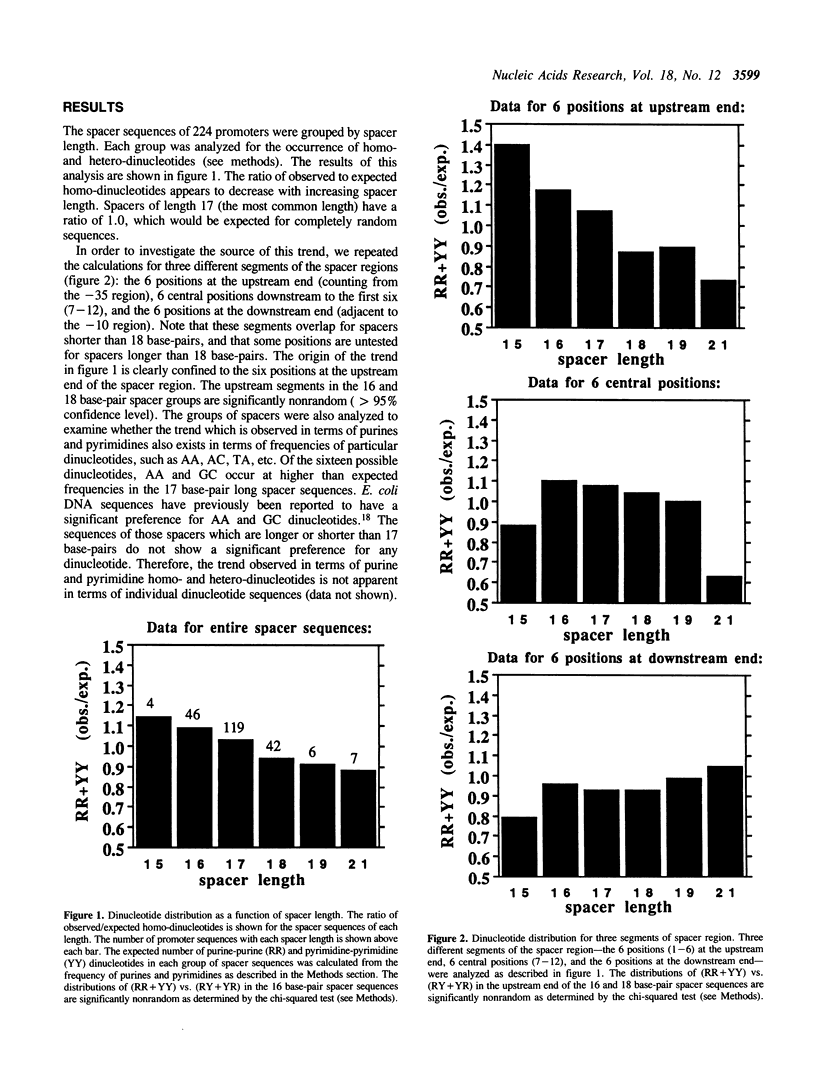
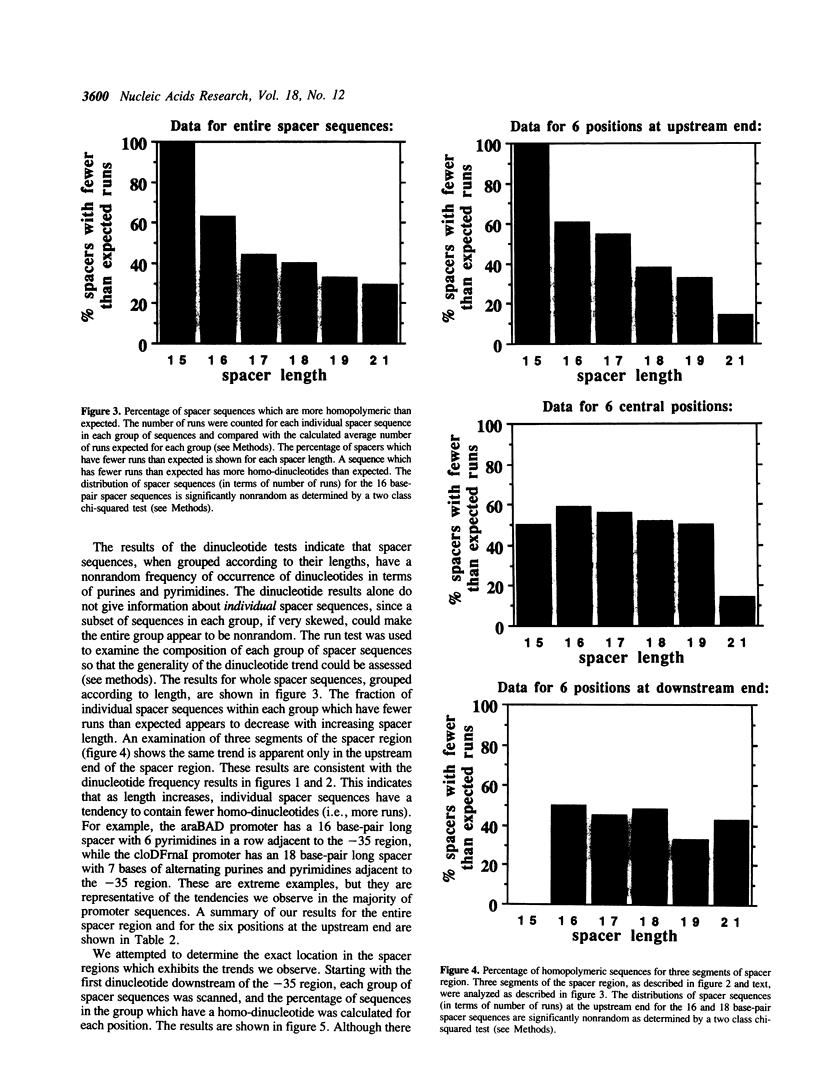
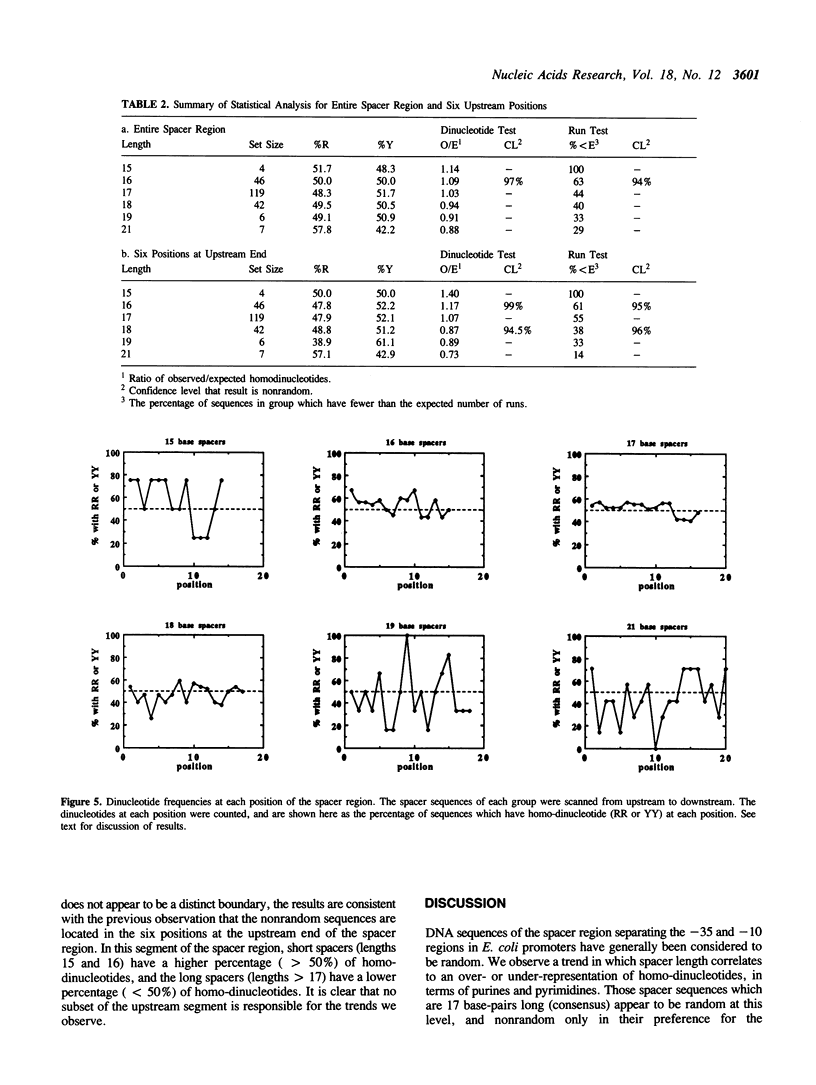
The -10 and -35 regions of E. coli promoter sequences are separated by a spacer region which has a consensus length of 17 base-pairs. This region is thought to contribute to promoter function by correctly positioning the two conserved regions. We have performed a statistical evaluation of 224 spacer sequences and found that spacers which deviate from the 17 base-pair consensus length have nonrandom sequences in their upstream ends. Spacer regions which are shorter than 17 base-pairs in length have a significantly higher than expected frequency of purine-purine and pyrimidine-pyrimidine homo-dinucleotides at the six upstream positions. Spacer regions which are longer than 17 base-pairs in length have a significantly higher than expected frequency of purine-pyrimidine and pyrimidine-purine hetero-dinucleotides at these positions. This suggests that the nature of the purine-pyrimidine sequence at the upstream end of spacer regions affect promoter function in a manner which is related to the spacer length. We examine the spacer sequences as a function of spacer length and discuss some possible explanations for the observed relationship between sequence and length.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auble D. T., Allen T. L., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Effects of substitutions in the spacer DNA separating the -10 and -35 regions. J Biol Chem. 1986 Aug 25;261(24):11202–11206. [PubMed] [Google Scholar]
- Ayers D. G., Auble D. T., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Role of the spacer DNA in functional complex formation. J Mol Biol. 1989 Jun 20;207(4):749–756. doi: 10.1016/0022-2836(89)90241-6. [DOI] [PubMed] [Google Scholar]
- Brosius J., Erfle M., Storella J. Spacing of the -10 and -35 regions in the tac promoter. Effect on its in vivo activity. J Biol Chem. 1985 Mar 25;260(6):3539–3541. [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Duval-Valentin G., Ehrlich R. Interaction between E. coli RNA polymerase and the tetR promoter from pSC101: homologies and differences with other E. coli promoter systems from close contact point studies. Nucleic Acids Res. 1986 Mar 11;14(5):1967–1983. doi: 10.1093/nar/14.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozinski T., Markiewicz W. T., Wyrzykiewicz T. K., Wierzchowski K. L. Effect of the sequence-dependent structure of the 17 bp AT spacer on the strength of consensuslike E.coli promoters in vivo. Nucleic Acids Res. 1989 May 25;17(10):3855–3863. doi: 10.1093/nar/17.10.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandecki W., Reznikoff W. S. A lac promoter with a changed distance between -10 and -35 regions. Nucleic Acids Res. 1982 Feb 11;10(3):903–912. doi: 10.1093/nar/10.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Brosius J., McClure W. R. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem. 1985 Mar 25;260(6):3529–3538. [PubMed] [Google Scholar]
- Nussinov R. Doublet frequencies in evolutionary distinct groups. Nucleic Acids Res. 1984 Feb 10;12(3):1749–1763. doi: 10.1093/nar/12.3.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M. C. Escherichia coli promoters. I. Consensus as it relates to spacing class, specificity, repeat substructure, and three-dimensional organization. J Biol Chem. 1989 Apr 5;264(10):5522–5530. [PubMed] [Google Scholar]
- Schroeder S. A., Roongta V., Fu J. M., Jones C. R., Gorenstein D. G. Sequence-dependent variations in the 31P NMR spectra and backbone torsional angles of wild-type and mutant Lac operator fragments. Biochemistry. 1989 Oct 17;28(21):8292–8303. doi: 10.1021/bi00447a006. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Stefano J. E., Gralla J. D. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Youderian P., Bouvier S., Susskind M. M. Sequence determinants of promoter activity. Cell. 1982 Oct;30(3):843–853. doi: 10.1016/0092-8674(82)90289-6. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]


