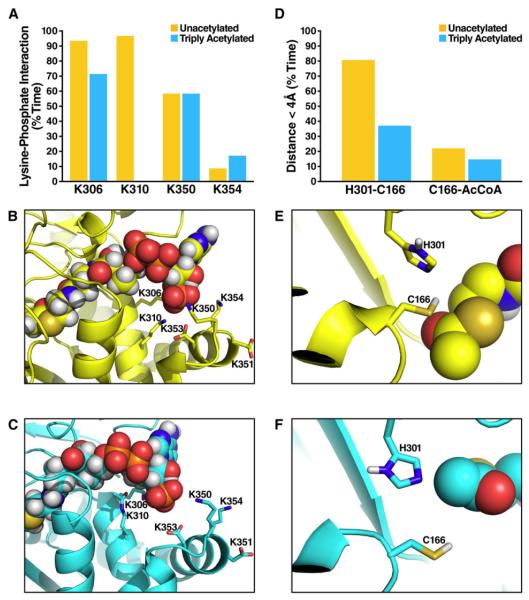
Figure 4. Acetylation Shifts Catalytic Residues in the Acetyl-CoA Binding Region.
(A) The percentage of time the distance between the 3′ phosphate atom of acetyl-CoA P(3′) and the nitrogen atom (N) in the sidechain of K310, K306, K350, and K354 is smaller than 4.2 Å, during which the phosphate group of acetyl-CoA and lysine sidechain form salt bridge (Mandell et al., 2007); comparing the unacetylated (yellow) and triply acetylated (cyan) HMGCS2; data extracted from the last 5 ns of each simulation and averaged over five independent simulations for either state of HMGCS2.
(B) Representative snapshot image during a simulation of the electrostatic network formed by K310, K306, K350, K354, and the 3′ phosphate group of acetyl-CoA in unacetylated HMGCS2.
(C) Same as (B) for the triply acetylated (K310, K447, K473) HMGCS2.
(D) The percentage of time the distance between the catalytic cysteine (C166) and nearby histidine (H301) (or acetyl-CoA) is smaller than 4.0 Å; comparing the unacetylated (yellow) and triply acetylated (cyan) HMGCS2; data extracted from the last 5 ns of each simulation and averaged over five independent simulations for either state of HMGCS2.
(E) Representative snapshot image during a simulation of the catalytic amino acids and the acetyl-CoA in unacetylated HMGCS2.
(F) Same as (E) for the triply acetylated (K310, K447, K473) HMGCS2.