Large quantities of recombinant human carboxylesterase 1 have been produced in an economical whole insect larvae system. The crystal structure of this enzyme is essentially identical to that produced by cell culture techniques.
Keywords: human carboxylesterase 1, whole-insect expression system, Trichoplusia ni
Abstract
The use of whole insect larvae as a source of recombinant proteins offers a more cost-effective method of producing large quantities of human proteins than conventional cell-culture approaches. Human carboxylesterase 1 has been produced in and isolated from whole Trichoplusia ni larvae. The recombinant protein was crystallized and its structure was solved to 2.2 Å resolution. The results indicate that the larvae-produced enzyme is essentially identical to that isolated from cultured Sf21 cells, supporting the use of this expression system to produce recombinant enzymes for crystallization studies.
1. Introduction
Human carboxylesterase 1 (hCES1) is expressed primarily in the liver, where its main function is assumed to be the metabolism of drugs and xenobiotics. This enzyme has the capacity to cleave ester, amide-ester and thioester bonds found in many xenobiotic compounds. hCES1 is a member of the serine esterase family that includes acetylcholinesterase (AChE) and butyrylcholinesterase (BuChE). Like AChE and BuChE, hCES1 is inhibited by organophosphorus compounds, including pesticides and chemical warfare nerve agents. The structure of hCES1 has been determined in covalent acyl-enzyme intermediate complexes with the nerve agents soman and tabun (Fleming et al., 2007 ▶) as well as cyclosarin (Hemmert et al., 2010 ▶). The hCES1 used in these previous studies was purified from the supernatant of Spodoptera frugiperda Sf21 cells infected with an appropriate baculovirus.
Insect cell lines derived from the cabbage looper moth Trichoplusia ni (T. ni) are often employed for protein expression in combination with baculoviral expression vectors. An alternative method for protein production is the utilization of whole insect larvae in conjunction with the Chesapeake PERL PERLXpress protein-expression platform (Welch et al., 2006 ▶; Otto et al., 2010 ▶). This approach is based on infection of T. ni larvae with an orally active baculovirus and results in the production of gram quantities of protein in a cost-efficient manner. The objective of this report was to assess the structural similarity of recombinant hCES1 from T. ni larvae to the same enzyme produced in other expression systems. Baculovirus encoding the complete amino-acid sequence of hCES1 was used to orally infect T. ni larvae in the fifth instar of development. After expression and purification, the protein was crystallized and its structure was solved. Despite the fact that the protein crystallized in a novel space group, the hCES1 structure reported here is essentially identical to that reported for hCES1 produced in Sf21 cell culture, thereby validating this method of protein production. The final coordinates and structure factors have been deposited in the Protein Data Bank (entry 4ab1). An Interactive 3D Complement (I3DC) page appears in Proteopedia for this study at http://proteopedia.org/w/Journal:Acta_Cryst_F:1.
2. Materials and methods
2.1. Expression and purification
Human carboxylesterase 1 (GenBank NM_001266.4) DNA was cloned into transfer vector pVL1393 and then incubated in the presence of linearized Autographa californica multiple nuclear polyhedrosis virus and Sf9 insect cells to form budded recombinant virus encoding the hCES1 protein (rhCES1). The hCES1 gene was expressed under the control of the polyhedron promoter and was engineered to contain an insect-derived N-terminal leader sequence. T. ni insect larvae were infected with recombinant virus by hand injection to form oral inocula expressing a pre-occluded form of the recombinant baculovirus (POV). Production larvae (∼1 kg) were prepared by oral inoculation using formulated POV applied to the insect diet followed by incubation for 96 h under controlled temperature and humidity. Larvae were harvested, frozen and stored at 193 K. For recovery, 40 g of recombinant insect larvae expressing rhCES1 were homogenized by gentle mechanical disruption using a tissue homogenizer. The homogenate was clarified by centrifugation followed by filtration using a 0.2 µm membrane. Viral inactivation was by Triton X-100 treatment followed by a second 0.2 µm filtration. The filtrate was bound to a Fractogel EMD SO3 (M) cation-exchange column (Merck, Darmstadt, Germany), washed and eluted. The material was desalted and then further purified by passage over a Toyopearl GigaCap Q-650 anion-exchange column (Tosoh Bioscience, King of Prussia, Pennsylvania, USA). The purified rhCES1 was formulated in 50 mM Tris pH 8.0, 75 mM NaCl. The protein concentration was determined using a bicinchoninic acid assay and quantified by a densitometric scan of a Coomassie-stained SDS–PAGE gel. The protein identity was confirmed by Western analysis using a protein-specific antibody (ab52941; Abcam, Cambridge, Massachusetts, USA). The total protein yield was 9 mg at 95% purity. The final protein concentration was 11 mg ml−1 and the sample was stored in a buffer consisting of 50 mM Tris pH 7.6, 150 mM NaCl. The protein was produced and purified by Chesapeake Protein Expression and Recovery Laboratories (Savage, Maryland, USA).
2.2. Functional analysis of rhCES1
70 ng rhCES1 was incubated with increasing concentrations of p-nitrophenyl butyrate (Sigma–Aldrich, St Louis, Missouri, USA) in 100 mM potassium phosphate buffer pH 7.0. The rate of formation of p-nitrophenol was followed at A 412 (∊ = 17 000 M −1 cm−1) with a SpectraMax Plus 384 spectrophotometer (Molecular Devices, Sunnyvale, California, USA) for 5 min at room temperature in a 96-well microplate.
2.3. Crystallization and data collection
Large single crystals grew in several conditions, but the most usable examples were obtained using a mixture consisting of 45% protein solution, 45% of condition No. 83 of the JCSG+ Suite (2 M ammonium sulfate, 0.1 M bis-tris pH 6.5, adjusted from pH 5.5) from Qiagen (Germantown, Maryland, USA) and 10% of condition No. 21 of the Additive Screen (2 M sodium thiocyanate) from Hampton Research (Aliso Viejo, California, USA) with a final volume of 800 nl. Drops were prepared in a Greiner (Frickenhausen, Germany) CrystalQuick LP 96-well sitting-drop plate using a Mosquito robot (TTP LabTech, Royston, UK) and stored at 292 K. A crystal was harvested from the drop and flash-cooled in liquid nitrogen for data collection at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Data-collection parameters are summarized in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest shell.
| Data collection | |
| ESRF beamline | ID23-2 |
| Wavelength (Å) | 0.8726 |
| Resolution range (Å) | 50–2.20 (2.24–2.20) |
| Space group | P6322 |
| Unit-cell parameters (Å) | a = b = 114.8, c = 177.1 |
| No. of unique reflections | 35656 (1744) |
| Overall Rmerge (%)† | 11.9 (65) |
| Percentage of reflections with I/σ(I) < 3.0 in highest resolution shell | 51.0 |
| Overall multiplicity | 16.8 (14.1) |
| Overall completion | 100 (99.8) |
| Refinement | |
| Resolution range (Å) | 47.9–2.20 |
| Rcryst‡ (working set) | 0.178 |
| Rfree§ | 0.236 |
| No. of protein atoms | 3980 |
| No. of water molecules | 241 |
| No. of heteroatoms | 23 |
| R.m.s.d. bond lengths (Å) | 0.020 |
| R.m.s.d. bond angles (°) | 1.98 |
| Mean B value (Å2) | 26.00 |
R
merge = 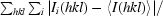
 , where I
i(hkl) and 〈I(hkl)〉 are the ith and the mean measurements of the intensity of reflection hkl, respectively.
, where I
i(hkl) and 〈I(hkl)〉 are the ith and the mean measurements of the intensity of reflection hkl, respectively.
R
cryst = 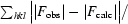
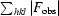 .
.
R free is equivalent to R cryst but calculated for a random subset (5.2%) of reflections that were omitted from the refinement process.
A search model based on a monomer of hCES1 (PDB entry 2hrq; Fleming et al., 2007 ▶) was used for molecular replacement with MOLREP (Vagin & Teplyakov, 2010 ▶), which is part of the CCP4 suite (Winn et al., 2011 ▶), which gave a clear solution. Small changes to the structure based on inspection of 2F o − F c and F o − F c maps in Coot (Emsley et al., 2010 ▶) were made over the course of nine cycles of refinement with REFMAC (Murshudov et al., 2011 ▶). The final model consisted of 524 residues, 241 water molecules, three thiocyanate ions and one N-acetylglucosamine residue bound to Asn79.
3. Results and discussion
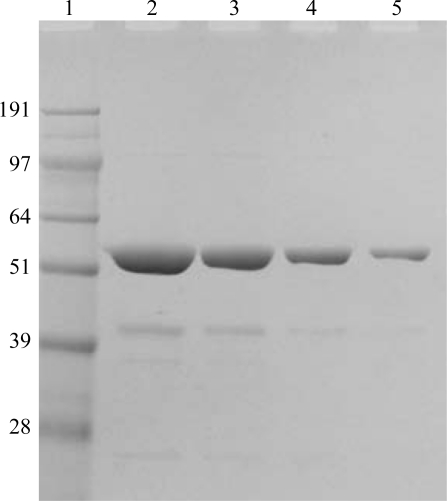
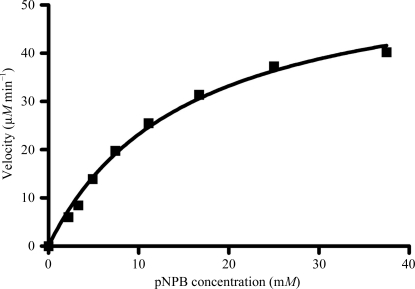
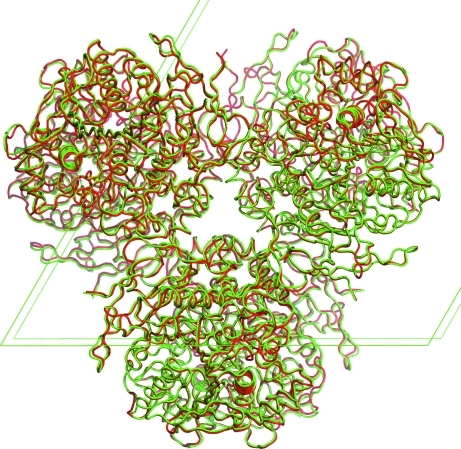
rhCES1 was expressed and purified from T. ni larvae. When the protein was analyzed by SDS–PAGE a 60 kDa band was observed (Fig. 1 ▶), which is in agreement with the theoretical value of 62 kDa for hCES1. The identity of the protein was confirmed by Western blot analysis using a CES1-specific antibody (data not shown). The protein hydrolyzed the substrate p-nitrophenyl butyrate (pNPB), demonstrating functional hCES1 enzymatic activity (Fig. 2 ▶). The current structure of rhCES1 represents the first published hexagonal crystal form, despite the fact that all other published examples of hCES1 structures consist of a hexamer in the asymmetric unit. The trimer of subunits sits around one of the threefold axes found in this space group, while the three twofold axes at z = 1/4 that intersect on this axis complete the hexamer. An alignment (LSQ superpose facility in Coot) of the A chain from PDB entry 2h7c (Bencharit et al., 2006 ▶) with the asymmetric unit reported here gave an r.m.s. deviation of 0.42 Å for 522 Cα atoms. An r.m.s. value of 0.47 Å (3132 Cα atoms) was obtained for the entire 2h7c hexamer superposed with the symmetry-generated rhCES1 hexamer, indicating that the quaternary structure is essentially identical (Fig. 3 ▶) in these crystal forms.
Figure 1.
SDS–PAGE analysis of rhCES1 expressed and purified from T. ni larvae. Various amounts of purified rhCES1 protein were separated on a 10% bis-tris polyacrylamide gel and visualized by Coomassie Blue staining. Lane 1, molecular-weight markers; lane 2, 8 µg rhCES1; lane 3, 4 µg rhCES1; lane 4, 2 µg rhCES1; lane 5, 1 µg rhCES1. Molecular-weight markers are labelled in kDa.
Figure 2.
Hydrolysis of p-nitrophenyl butyrate (pNPB) by rhCES1. A velocity versus concentration plot for rhCES1 with pNPB is shown.
Figure 3.
Overlay of the hexameric asymmetric unit of 2h7c (red Cα trace) with the hexamer of rhCES1 (green). The fit is based on the structural overlap of one chain of 2h7c with the asymmetric unit of rhCES1. Unit-cell edges are shown as green lines. This figure was prepared with PyMOL v.1.1r1 (DeLano, 2002 ▶).
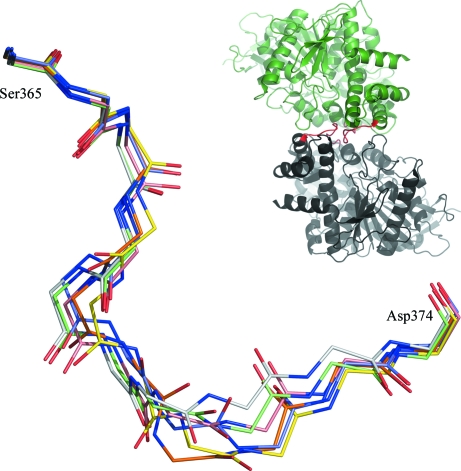
Regions of the current structure that differ from the previously reported examples of hCES1 include Ala338–Thr342, which has little density, and Ser365–Asp374, which has very poor density. All six examples of the former region in the monoclinic structure 2hrq have the highest B factors in their respective structures. While 2h7c is also monoclinic and has a hexamer in the asymmetric unit, the molecules pack differently with respect to 2hrq. Thus, in 2h7c the region Ala338–Thr342 forms some close crystal contacts, especially in the case of chain A, giving lower temperature factors and improved electron density for some instances of this region in the hexamer.
In the 2h7c structure each of the six examples of the section of chain from Ser365 to Asp374 has a different conformation, as shown in Fig. 4 ▶. The poorly defined density for this same region in rhCES1 is consistent with the observation that this loop can adopt multiple conformations despite its participation in forming the twofold interface between chains (see inset in Fig. 4 ▶).
Figure 4.
Overlay of all six copies of the region from Ser365 to Asp374 in 2h7c, showing that each copy follows a unique trace. The inset shows two representative chains from 2h7c, with the region in question coloured red. This figure was prepared with PyMOL.
The current results confirm that rhHCES1 isolated from the T. ni system is essentially identical to previous examples of this enzyme isolated from cultured insect cells, validating the use of the whole-insect system as a source for recombinant proteins in structure-determination studies.
Supplementary Material
PDB reference: human carboxylesterase 1, 4ab1
Acknowledgments
We thank the Israel Structural Proteomics Centre for access to their facilities and Robert Balcerzak for assistance. We gratefully acknowledge financial support from the Defense Threat Reduction Agency (DTRA) Joint Science and Technology Office, Medical S&T Division (Projects HDTRA1-11-C-0026 and CBM.SCAV.01.10.RC.017). JLS is the Pickman Professor of Structural Biology. The views expressed in this manuscript are those of the author(s) and do not reflect the official policy of the Department of Army, Department of Defense, or the US Government.
References
- Bencharit, S., Edwards, C. C., Morton, C. L., Howard-Williams, E. L., Kuhn, P., Potter, P. M. & Redinbo, M. R. (2006). J. Mol. Biol. 363, 201–214. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fleming, C. D., Edwards, C. C., Kirby, S. D., Maxwell, D. M., Potter, P. M., Cerasoli, D. M. & Redinbo, M. R. (2007). Biochemistry, 46, 5063–5071. [DOI] [PMC free article] [PubMed]
- Hemmert, A. C., Otto, T. C., Wierdl, M., Edwards, C. C., Fleming, C. D., MacDonald, M., Cashman, J. R., Potter, P. M., Cerasoli, D. M. & Redinbo, M. R. (2010). Mol. Pharmacol. 77, 508–516. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otto, T. C., Kasten, S. A., Kovaleva, E., Liu, Z., Buchman, G., Tolosa, M., Davis, D., Smith, J. R., Balcerzak, R., Lenz, D. E. & Cerasoli, D. M. (2010). Chem. Biol. Interact. 187, 388–392. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Welch, R. W. et al. (2006). Bioprocess. J. 5, 63–70.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human carboxylesterase 1, 4ab1