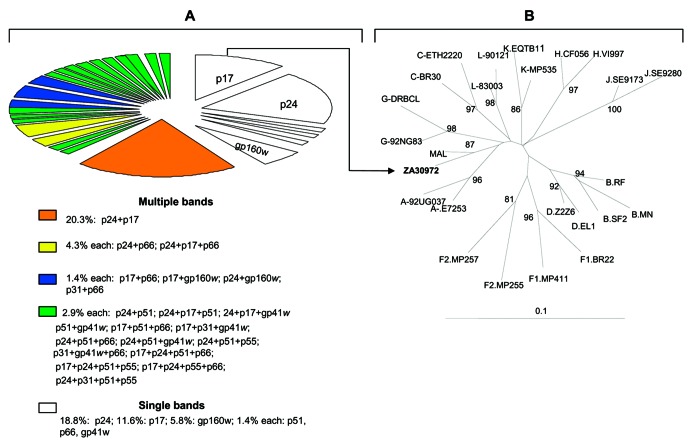
Figure.
A) Distribution of HIV-1 Western blot–indeterminate patterns among 69 serum specimens from Kinshasa, Zaire, reactive by enzyme immunoassay. B) Phylogenetic classification of HIV-1 protease sequence ZA30972 (GenBank accession no. AY562558) isolated from the p17 gag-reactive serum. The phylogenetic tree was generated by the neighbor-joining method with the nucleotide distance calculated by Kimura 2-parameter method (12), included in the GeneStudio package (http://www.genestudio.com). Reference sequences were obtained from the Los Alamos database (http://hiv-web.lanl.gov/MAP/hivmap.html). The position of the outgroup (Simian immunodeficiency virus [SIV]cpz) is not shown. Values on branches represent the percentage of 100 bootstrap replicates. The scale bar indicates an evolutionary distance of 0.10 nucleotides per position in the sequence.