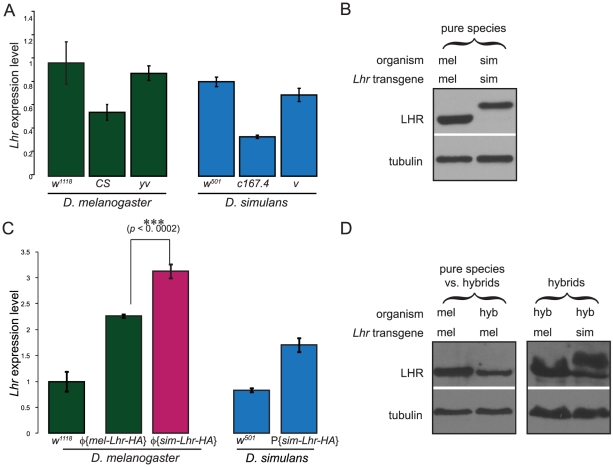
Figure 5. Cis-by-trans regulatory divergence of D. simulans Lhr.
(A) Lhr transcript levels in different wild type and marker strains of D. melanogaster and D. simulans (nested ANOVA F1;4 = 0.89; p = 0.39 for between species variation). (B) LHR protein levels are similar between D. melanogaster (mel) and D. simulans (sim). Western blot detecting HA-tagged LHR transgenes expressing in their own species. sim-LHR-HA migrates higher than mel-LHR-HA. (C) Lhr transcript levels in transgenic lines compared to the corresponding host strain genetic background (w1118 for D. melanogaster and w501 for D. simulans). The transgenic lines are homozygous for the transgene and for the endogenous Lhr allele, and therefore have four copies of Lhr. Lhr transcript abundance is approximately doubled in φ{mel-Lhr-HA} and P{sim-Lhr-HA} relative to their respective reference backgrounds, indicating wild type expression levels of these transgenes in their native species. In contrast, Lhr transcription in φ{sim-Lhr-HA} is significantly higher than in φ{mel-Lhr-HA} (by two-tailed t-test) and is ∼3× the level of the reference background. (D) LHR protein levels are not increased in hybrids. Left, one copy of the mel-Lhr-HA transgene in D. melanogaster and in hybrids; right, one copy of the mel-Lhr-HA or sim-Lhr-HA transgenes in hybrids. The origin of the lower band in the sim-Lhr-HA lane in hybrids is unclear. For A and C, RNA was isolated from 6–10 hr old embryos. Lhr expression levels were measured relative to rpl32 using quantitative RT-PCR. Expression levels were normalized by setting the D. melanogaster w1118 strain to 1. Error bars represent standard deviation within biological replicates, n≥6 for all except P{sim-Lhr-HA} where n = 3. For B and D, protein extracts were from 0–16 hr embryos and the immunoblots were hybridized with anti-HA antibodies. Anti-tubulin antibodies were used as a loading control.