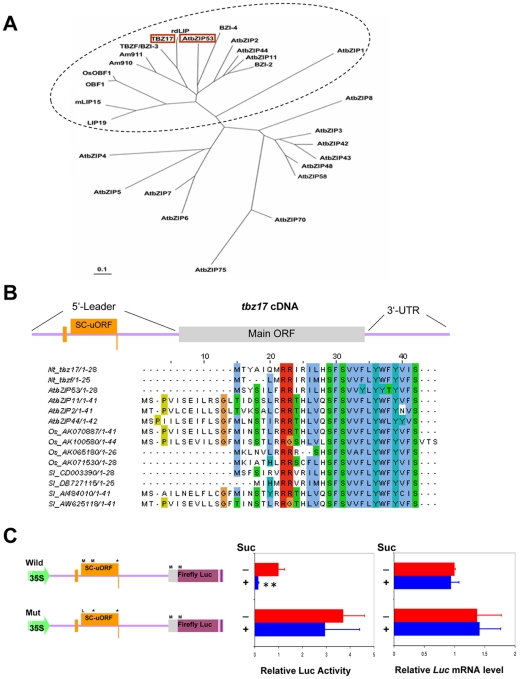
Figure 1. SIRT is found in tobacco tbz17 gene.
A, Phylogenetic relationship between 17 bZIP proteins of Arabidopsis class S [7] and LIP19 subfamily members including tobacco TBZ17. LIP19 subfamily is indicated by dotted-circle line and TBZ17 and AtbZIP53 are highlighted. The amino acid sequence alignment was constructed by the ClustalW program and the relationship was visualized by TREEVIEW program [39]. AtbZIP1 (At5g49450), AtbZIP2 (GBF5, At2g18160), AtbZIP11 (ATB2, At4g34590), AtbZIP44 (At1g75390), AtbZIP53 (At3g62420), Am910 (Y13675), Am911 (Y13676), BZI-2 (AY045570), BZI-4 (AY045572), LIP19 (X57325), mLIP15 (D26563), OBF1 (X62745), rdLIP (AB015187), TBZ17 (D63951), TBZF (identical to BZI-3, AB032478), OsOBF1 (AB185280). B, Structure of tobacco tbz17 cDNA and its evolutionary conserved uORF. The first, second and third uORFs in the 5′-leader region are positioned in the second-, first- and third-frames of tbz17 cDNA, respectively. The second uORF, here called SC-uORF, is highly conserved. The predicted amino acid sequence encoded by tbz17 SC-uORF is aligned with those of the other group S bZIP-encoded cDNAs. Nt, Nicotiana tabacum; At, Arabidopsis thaliana; Os, Oryza sativa; Sl, Solanum lycopersicum. C, SIRT found in tbz17 is mediated by its SC-uORF. The constructs used are as follows; Wild: the intact tbz17 5′-leader sequence (+1 to +358 of tbz17 cDNA) was inserted between the CaMV 35S promoter and the firefly luciferase (LUC) gene. Mut: the start codon (ATG) of SC-uORF is mutated to Leu codon (TTG) and the second Met codon (ATG) is replaced by stop codon (TAA) by site-directed mutagenesis. The constructs were delivered into Arabidopsis thaliana rosette leaves (collected from 3 week-old-seedlings). The bombarded leaves were incubated in half-strength MS media for 20 h with or without 6% sucrose. The relative LUC mRNA levels and LUC activities were analyzed. The error bar represents the SD. Asterisk indicates significant differences that were observed due to sucrose treatment (Student’s t-test: **P<0.01).