Figure 2.
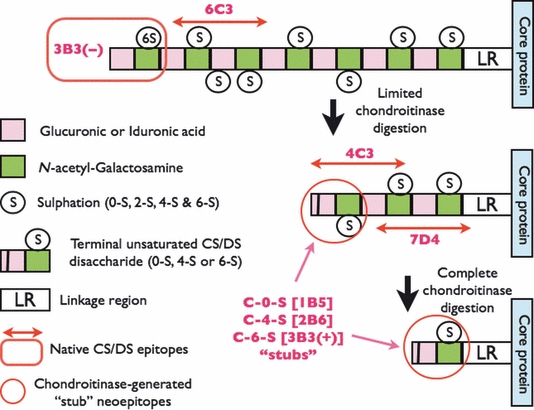
Structure and location of native epitopes [for mAbs 3-B-3(−), 4-C-3, 6-C-3 and 7-D-4] and chondroitinase-generated ‘stub’ neoepitopes [for mAbs 1-B-5, 2-B-6 and 3-B-3(+)] on chondroitin sulphate (CS) and dermatan sulphate (DS) glycosaminoglycan chains. Boxed in Red is the location of mAb 3-B-3(−) native epitope that recognizes a non-reducing terminal end saturated CS disaccharide consisting of glucuronic acid that is adjacent to N-acetylgalactosamine-6-sulphate. The native epitope for mAb 6-C-3 (Red linear arrows) is located in CS/DS oligosaccharide sequences that are located on the outer peripheral regions of the CS/DS GAG chains whilst mAbs 4-C-3 and 7-D-4 are located in oligosaccharide sequences located in the inner regions of the CS/DS GAG chains nearer to the linkage region (LR) that is the covalent attachment site that links CS/DS GAGs to the proteoglycan core protein. The chondroitinase-generated neoepitope ‘CS stubs’ are circled in Red and occur as an unsaturated uronic acid residue that is adjacent to N-acetylgalactosamine that is either non-sulphated (C-O-S; recognized by mAb 1-B-5), 4-sulphated (C-4-S; recognized by mAb 2-B-6) or 6-sulphated [C-6-S; recognized by mAb 3-B-3(+)]. CS/DS GAG chains are depicted as a sequence of disaccharides with two different coloured boxes that have either been partially (limited) or completely digested (deglycosylated) with chondroitinase. Chodroitinases are enzymes that remove CS/DS disaccharides with non-reducing unsaturated uronic acid residues from the CS/DS GAG chains eventually leaving a characteristic terminal disaccharide ‘stub’, that is, either unsulphated, 4- or 6-sulphated, and covalently attached to the proteoglycan core protein.
