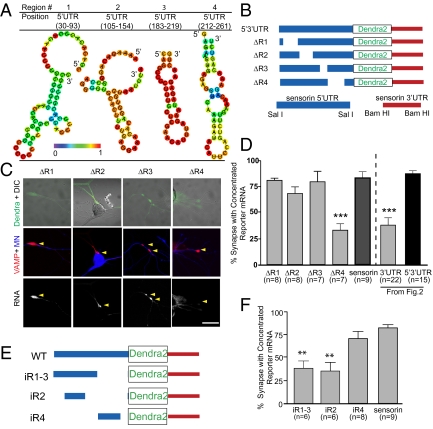
Fig. 3.
A region directly upstream of the sensorin translation start site, in the 5′ UTR, is necessary and sufficient for synaptic localization of the reporter mRNA. (A) Representative graphic depictions of potential stem-loop structures in the 5′ UTR of sensorin predicted by RNAfold (35); color scale denotes base pair probabilities. (B) Cartoon representation of deletions made to the reporter construct. (C) Representative images of deletion constructs ΔR1–4 expressed in SNs synaptically connected with MNs. Top: Photomicrograph of Dendra protein (SN, green) merged with DIC image. Middle: Synapses marked as VAMP-mCherry clusters (red) contacting the MN (blue, Alexa 647). Bottom: FISH images showing clustering of reporter mRNA. (D) Quantification of synaptic localization of reporter mRNAs as the percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA. (E) Cartoon of insertion constructs (iR1–4) in which the designated regions of the sensorin 5′ UTR were inserted into a control pNEX 5′ UTR to test their ability to localize the reporter mRNA to synapses. The mutants were coexpressed in SNs paired with target MNs at DIV 2, and neurons were fixed on DIV 4 and processed for FISH. (F) Percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA.