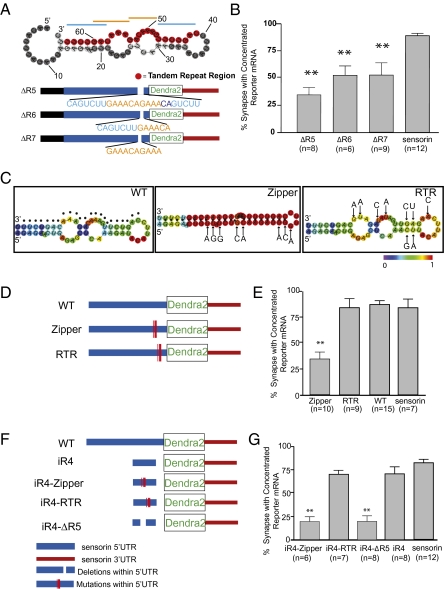
Fig. 4.
Stem-loop structure (66 nt) localizes reporter mRNA to synapses. (A) Predicted secondary structure of the region corresponding to R4 [RNAfold (35); highlighted in red is the 24-nt primary sequence containing a double-tandem 7mer-repeat (indicated with blue/orange lines)]. Mutants were generated in which the entire 24-nt (ΔR5), the first 13-nt set of repeat elements (ΔR6), or 10 nt of the center repeat (ΔR7) were deleted from the 5′3′ UTR reporter construct. (B) Mutants were coexpressed with VAMP-mCherry in SNs paired with target MNs, and the percentage of synapses containing reporter RNA was measured. **P < 0.01, one-way ANOVA followed by Dunnett's multiple comparison test compared with 3′ UTR reporter or endogenous sensorin. See also Figs. S2, S3, and S5. (C) Predicted secondary structures [RNAfold (35)] of WT (dots denote tandem repeat region described in Fig. 3); zipper construct, 8-point mutations were introduced to collapse the predicted secondary structure without disrupting the tandem repeat sequence; RTR construct, 9 mutations were introduced to disrupt the primary sequence of the tandem repeat region, while retaining predicted secondary structure. (D) Cartoon showing the location within the sensorin 5′ UTR of the mutations in the zipper and RTR constructs. The mutants were coexpressed in SNs paired with target MNs at DIV 2, and neurons were fixed on DIV 4 and processed for FISH. (E) Percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA. (F) Cartoon of mutant insertion reporter constructs, including WT iR4 (66 nt), iR4-RTR (iR4 with mutations shown in RTR in C), iR4-Zipper (iR4 with mutations shown in zipper in C), or iR4-ΔR5 (iR4 lacking the 24-nt repeat element). The mutants were coexpressed in SNs paired with target MNs at DIV 2, and neurons were fixed on DIV 4 and processed for FISH. (G) Percentage of synapses (VAMP-mCherry clusters adjacent to MN) containing reporter mRNA. **P < 0.001, one-way ANOVA followed by Dunnett's multiple comparison test.