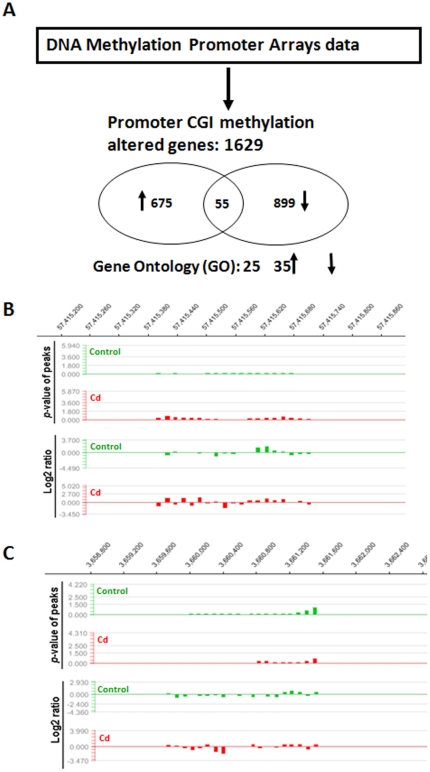
Figure 2. Effects of chronic exposure to Cd to genomic DNA methylation in rat livers.
DNA methylation in the livers of rats with and without low-dose Cd treatment was initially profiled with MeDIP-chip, followed by GO analysis as described in Materials and Methods . The brief summary of these analyses is outlined (A) with the numbers of total genes with promoter CGI methylation changes (1,629) and the genes, for which all CGIs in the promoter were either hypermethylated (675) or hypomethylated (899). In addition, there were also some genes (55) with mixed hyper- and hypo-methylated CGIs in each promoter. After general analysis for the DNA methylation changes in the livers of rats with and without low-dose Cd treatment as described in A, the methylation profiles of the promoter CGI of CASP8 (B) and TNF (C) genes were selectively analyzed. The NimbleScan software was employed based on the raw data described in Table S1, to generate scaled log2-ratio data for IP/input values and p-value enrichment data for each probe. Peaks (methylated regions) are then generated based on the p-value data. Therefore, the methylation signal levels (Y axis) are represented as either p-value of peaks (top track in each panel) or log2-ratio (bottom track in each panel) along with different regions of the promoter gene. For each gene there are only positive numbers relative to base line (0) in term of p-value of peaks for each gene, but there are both negative and positive numbers for log2-ratio. Conclusion was made by comparing the overall values between control and Cd-treated groups.