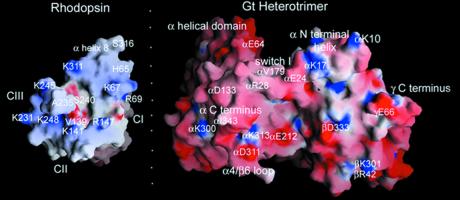
Figure 2.
grasp (http://trantor.bioc.columbia.edu/grasp) views of the interacting surfaces between rhodopsin's cytoplasmic face and Gt's rhodopsin-interacting surface. Imaginary folding on the dotted line will dock the two molecules in the “best guess” complementary surface. The cytoplasmic face of rhodopsin is relatively small and has a distinct orientation, because rhodopsin is a transmembrane protein. Coordinates from the refined rhodopsin structure [Protein Data Bank (PDB) no. 1HZX (37)] and Gt [PDB entry 1GOT (23)] were used in grasp to examine complementary surfaces of the two molecules. The extreme carboxyl-terminal residues of rhodopsin after S316 are not involved in G protein binding and occlude the intracellular loops. These residues were removed from the grasp view for clarity. This is the ground state of rhodopsin, and critical activating residues such as the ERY sequence are buried in the structure. The loops making up the cytoplasmic face are somewhat disordered in the crystal structure. Still, there is an overall complementarity of shape between the sites of interaction that might already be used to guide mutagenesis on Gα. Notice the overall charge complementarity and the more explicit charge complementarity between residues K341, K248, K141, and R147 on rhodopsin and D311 and E212 on Gα at the bottom of both molecules. Also notice that the deep pockets made up of the interhelical space in rhodopsin (where the flexible carboxyl-terminal residues from Gα could bind?) and the βγ cleft (where CIII residues could fit?) may be complementary.