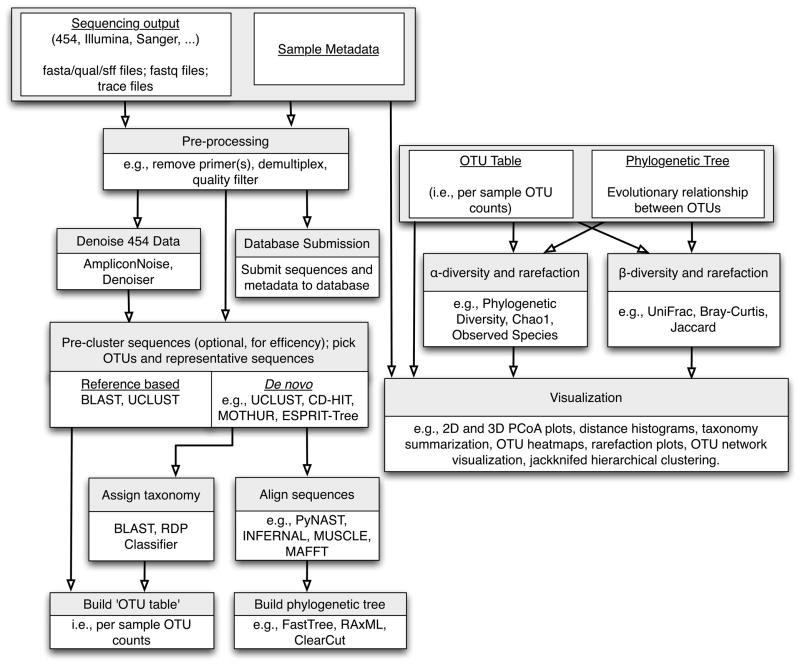
Figure 2.
High-throughput studies follow a common workflow that begins with raw sequence data and sample metadata (primer barcodes and environmental data). Raw data is filtered and processed, with the option of denoising (a step currently applicable only to 454 data) before Operational Taxonomic Units (OTUs) are picked through reference-based or de novo approaches. OTU picking can include pre-clustering steps such as Single Linkage Preclustering (SLP, [94]), prefix-suffix filtering or collapsing of identical sequences to reduce compute time (all methods available within the QIIME pipeline [51]); the recommended and default OTU picking workflow in QIIME currently involves sorting sequences by abundance, collapsing identical reads, picking OTUs de novo with uclust, and subsequently inflating the ‘identical reads’ to recapture abundance information about the initial sequences). Taxonomy is next assigned to OTU reference sequences, followed by construction of an OTU abundance matrix and a phylogenetic tree; when working with a closed reference-based OTU picking protocol it is not necessary to make taxonomic assignments or build a phylogenetic tree as these can be obtained directly from the reference data set. These outputs can be subsequently utilized for ecological diversity analyses and visualization approaches.