Abstract
We have reported isolation and characterization of the prostate-specific and androgen-regulated PrLZ gene abnormally expressed in prostate cancer. PrLZ is a potential biomarker for prostate cancer and a candidate oncogene promoting cell proliferation and survival in prostate cancer cells. A full delineation of the PrLZ gene and its gene products may provide clues to the mechanisms regulating its expression and function. In this report, we identified three additional exons in the PrLZ gene and recognized five transcript variants from alternative splicing that could be detected by RT-PCR and western blotting. Structural comparison demonstrated that the PrLZ proteins are highly conserved among species. PrLZ contains multiple potential sites for interaction with other proteins. We used mammalian two-hybrid assays to demonstrate that PrLZ isoforms interact with 14-3-3 proteins, and multiple sites in the PrLZ may be involved in the interaction. Alternative splicing may contribute to abnormally enhanced PrLZ levels in prostate cancer, and interaction with 14-3-3 proteins may be a mechanism by which PrLZ promotes cell proliferation and survival during prostate cancer development and progression. This information is a valuable addition to the investigation of the oncogenic properties of the PrLZ gene.
Keywords: PrLZ gene, prostate-specific gene, alternative splicing, protein interaction, 14-3-3, mammalian two-hybrid assay
Introduction
Prostate-specific genes are highly relevant to prostate cancer diagnosis and treatment. Proteins specific to the prostate may be used as markers for cancer detection, since prostate cancer development and progression is often accompanied by aberrant gene expression [1; 2; 3]. Aberrant gene expression is a contributing factor to prostate cancer. Functional studies have identified prostate-specific genes that provide cancer cells with growth, survival, and metastasis advantages [4; 5; 6]. The identification and characterization of prostate-specific genes will help elucidate the molecular mechanisms by which cancer cells progress. In addition, regulatory mechanisms controlling prostate specificity could be adapted for therapeutic targeting [7; 8].
We reported the isolation and characterization of a prostate-specific and androgen-regulated PrLZ gene [9]. This gene is located in chromosome 8q21.1, one of the most frequently amplified loci in prostate cancer [10], encoding a leucine zipper protein of the Tumor Protein D52 (TPD52) family [9]. Expression of the PrLZ gene is developmentally controlled [5], with the protein level correlated to proliferative activities of the prostate epithelial cells in early embryonic morphogenesis and young adult life. In later life, expression in normal prostate is declined to a minimal level. Expression of PrLZ is abnormally activated in prostate cancer [5; 9]. In clinical specimens, high levels of PrLZ are detected specifically in malignant cells in prostatic intraepithelial neoplasia, primary tumors, and bone metastasis [9], and the level of abnormal expression is positively correlated to tumor stage [5]. PrLZ is a potential biomarker for prostate cancer. Moreover, PrLZ is a candidate oncogene for prostate cancer development and progression, since overexpressing PrLZ promotes cancer cell proliferation and facilitates cell survival in vitro, and accelerates xenograft tumor growth in athymic mice [5].
Although PrLZ is known to be regulated developmentally and by androgens, the underlying regulatory mechanism is currently unclear. The cause of the abnormal activation in prostate cancer and the molecular mechanism by which PrLZ promotes cell growth and survival have yet to be elucidated. Detailed characterization of the PrLZ gene is vital for an in depth mechanistic inveatigation. In this report, we performed a structural characterization of the PrLZ gene and identified isoforms of the PrLZ protein rising from alternative splicing. By identifying three additional coding exons and five alternatively spliced isoforms, this work lays the foundation for mechanistic studies of the PrLZ gene.
Materials and Methods
Cell culture
The source of LNCaP, C4-2, C4-2B, PC3, DU145, and ARCaP human prostate cancer cell lines used in this study was previously reported [9]. Cells were maintained in T-medium (Invitrogen, Carlsbad, CA) with 5% fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100μg/ml) at 37° C with 5% CO2 in a humidified incubator.
Screening cDNA library and DNA sequencing analysis
Construction and screening of the ZAPExpress cDNA library of C4-2 human prostate cancer cells was described previously [9]. To isolate PrLZ cDNA clones, a 502 base pair (bp) cDNA fragment containing gene-specific region of the PrLZ was used as probe. Positive phage clones were rescued into pBK-CMV plasmid by in vitro autoexcision. Each cDNA clone was subjected to DNA sequencing analysis with PrLZ-specific primers 5’-ATGGATTGTAGAGAGATGGACTTATATGAG-3’ (forward) and 5’-TCACAGGCTCTCCTGTGTCTTTTCTG-3’ (reverse).
Structural comparison
Following a homology search through GanBank, additional sequences were retrieved and used in the alignment. These included rat PrLZ (GenBank accession numbers XM_215524), mouse PrLZ (AY048852), human TPD52 (NM_005079), mouse TPD52 (NM_009412), rabbit CSPP28 (U35428), frog PrLZ (BC075131), quail R10 (Y07757), and chicken R10 (AJ721090).
RT-PCR analysis
The protocol for RT-PCR and the reaction conditions were previously reported [9]. To compare gene expression in human prostate cancer cell lines, total RNA of a normal human prostate was purchased from Clontech (Mountain View, CA). To clone rat PrLZ, total RNA from Wistar rat prostate was used in first-strand cDNA synthesis. PCR primers 5’-ATGGATTGTAGAGATATGGAGCTGTCTGATG-3’ and 5’-TCAGGGGCTCTCTGTCGTCTGTTCTGGAGGTGGCTC-3’ were used. The product was cloned to pGEM-T Easy vector (Promega, Madison, WI) and analyzed by DNA sequencing. Other PCR primers used in the study are listed in Supplemental Data.
Mammalian two-hybrid assay
The CheckMate Mammalian Two-Hybrid System (Promega) was used to assay interaction between PrLZ and 14-3-3 proteins, employing the manufacturer’s recommended protocol. Full coding sequences for PrLZ and 14-3-3 were obtained from C4-2 cells by RT-PCR cloning and confirmed by DNA sequencing. The cDNA was then cloned in frame to the pACT and pBIND mammalian two-hybrid vectors. Each PrLZ construct in pACT was evaluated for interaction with each 14-3-3 in pBIND by co-transfecting the combined plasmids together with the pG5-luc luciferase reporter to C4-2 cells in a 6-cm culture dish. After 48 hours culture, cells were plated onto 6-well plates in triplicate. Cells were harvested for measuring luciferase activity after 72 hours of culture. In complementary experiments, PrLZ in pBIND was evaluated for interaction with 14-3-3 in pACT.
Western blotting
The western blotting protocol was reported previously [9]. Briefly, an equal amount of whole cell lysate protein (40 μg) was fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto nitrocellulose membrane. Production and characterization of the antibodies to PrLZ were reported previously [9].
Results
1. Isolation and characterization of alternatively spliced PrLZ isoforms
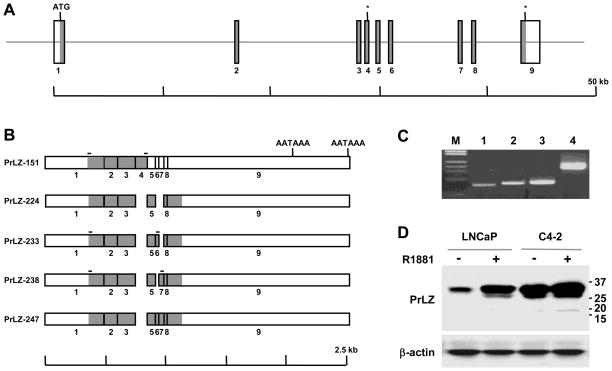
As previously reported, we isolated 24 full-length PrLZ clones by screening a C4-2 cDNA library [9]. In the present study, we further characterized these clones by determining DNA sequences with two primers flanking the coding region. This analysis revealed that while 15 of the clones contained coding sequences identical to our previously reported sequence [9], 3 other clones contained a 70 bp insert in the 5’ half of the coding sequence. Another clone contained a 27 bp insert, and two were found to contain a 42 bp insert in the 3’ half of the coding region. The other 3 clones contained both the 27 bp and 42 bp inserts in tandem. The DNA sequencing results thus revealed that the PrLZ gene in C4-2 cells produced five types of transcript variants.
We have isolated and characterized a genomic clone containing the PrLZ gene [9]. The genomic sequence of the gene is known. DNA sequence comparison confirmed that sequences for all the three inserts in the cDNA clones were present in the genomic clone as individual coding exons, increasing the number of exons in the PrLZ gene from 6 to 9 (Figure 1A). The 70 bp insert was from newly identified exon 4. The 27 bp insert found in the cDNA clones was from newly identified exon 6, and the 42 bp insert was from exon 7 (Figure 1B). Cloning and DNA sequencing studies thus revealed that the PrLZ gene contains 9 exons spanning a 44 kilobase genomic region (Figure 1A). Three of the newly identified exons (exons 4, 6, and 7) could be alternatively spliced, and alternative splicing yielded 5 transcript variants, which could be translated to five PrLZ protein isoforms (Figure 1B). In this report, the isoforms are designated PrLZ-151, PrLZ-224, PrLZ-233, PrLZ-238, and PrLZ-247, according to the number of amino acid residues (aa) contained in the isoforms. GenBank Accession numbers for the isoforms are GQ499324 (PrLZ-151), GQ499325 (PrLZ-224), GQ499326 (PrLZ-233), GQ499327 (PrLZ-238), and GQ499328 (PrLZ-247).
Figure 1. Structure of the PrLZ gene and alternatively spliced isoforms.
A, schematic presentation of PrLZ gene organization. Exons are shown as vertical bars and coding regions are shaded in gray. The translation start site is marked (ATG) and two in frame stop codons are marked with asterisks (*). Note that sizes of the exons are not in scale. B, schematic presentation of the PrLZ cDNA sequences due to alternative splicing. Two polyadenylation signals (AATAAA) are marked. On top of each scheme the location of primers used for RT-PCR detection of specific transcripts is marked with a horizontal bar. C, RT-PCR detection of the alternatively spliced transcripts in C4-2 prostate cancer cells. Specific transcripts detected are PrLZ-151 (lane 1), PrLZ 233 (lane 2), and PrLZ-238 (lane 3). GAPDH was used as control (lane 4). Commercial λHindIII/ΦX174HaeIII was used as a DNA marker (M). D, Western blot detection of PrLZ isoforms. In the upper panel, whole cell lysates (40 μg) of LNCaP and C4-2 cells treated with R1881 (+) were subjected to western blotting for PrLZ. Sizes (kD) of the bands were indicated on the right. The result is representative of two separate experiments. In the lower panel, the level of the β-actin was used as a loading control.
2. Detection of the PrLZ isoforms
To assess whether the PrLZ isoforms could be detected as transcripts, we performed RT-PCR analysis using isoform-specific primers to amplify variants of the PrLZ mRNA from the C4-2 cells, which expressed an elevated level of PrLZ [5; 9]. As shown in Figure 1C, RT-PCR with primers from exon 1 and the newly identified exon 4 detected a product (lane 1) with the expected coding sequence size from exon 1 to exon 4 of 456 bp. Using this method, we detected the variant transcripts containing exons 6 and 7, and products were obtained with the expected sizes of 533 bp and 575 bp respectively (lanes 2 and 3).
To detect the PrLZ isoforms at the protein level, we used 40 μg whole cell lysate proteins in western blotting with a prolonged exposure time (Figure 1D). Multiple bands of PrLZ were seen both in LNCaP cells treated with androgen and in C4-2 cells. In the sample where the C4-2 cells were treated with androgen, a specific band about 17 kD could be seen, most likely representing protein of the truncated PrLZ-151 isoform. These results demonstrated that PrLZ isoforms could be detected in the C4-2 cells, although the level of the PrLZ-151 isoform is low in this cell line.
3. Structural conservation of the PrLZ protein
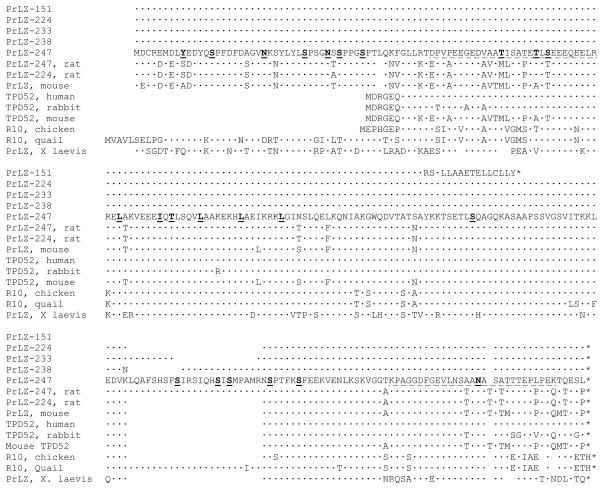
We performed a structural comparison of the PrLZ isoforms. Compared to the longest PrLZ-247 isoform, PrLZ-151 contained only the N-terminal half of the PrLZ (Figure 2). This was due to the inclusion of exon 4, which contained a stop codon in frame with the PrLZ coding sequence. Inclusion of this exon in mature mRNA resulted in a PrLZ isoform with a 16 aa leucine-rich tail not found in other isoforms, followed by a premature termination of the protein (Figure 2). The structure of PrLZ-224 has been reported. Compared to PrLZ-247, PrLZ-224 lacked a 23 aa domain in the C-terminal region, due to splicing of exons 6 and 7 (Figure 1B). In the same region, PrLZ-233 was found with a 9 aa insert encoded by exon 6, while PrLZ-238 contained a 14 aa sequence encoded by exon 7 (Figure 1B). In the PrLZ-238 isoform, the joint between exon 5 and exon 7 resulted in a codon change (from AAA to AAT), yielding a non-conservative replacement at the junction (Figure 2).
Figure 2. Amino acid sequences of the PrLZ isoforms and comparison with other TPD52 family members.
The upper five lines show structural differences between the PrLZ isoforms. The lower nine lines sequence conservation among TPD52 family members. Compared to PrLZ-247, an identical residue is marked with a dot. A gap is introduced to ensure the comparison reaches maximal homology. Along the PrLZ-247 sequence, the two PEST domains are underlined and the leucine zipper domain is marked by underlining the repetitive leucine (L)/isoleucine (I) residues in bold. Also underlined and in bold are consensus sites for tyrosine (Y) phosphorylation, serine (S)/threonine (T) phosphorylation, and N-glycosylation (N).
PrLZ proteins were small acidic polypeptides. As previously reported, migration of the PrLZ isoforms in SDS-PAGE was retarded, probably due to protein modifications including phosphorylation and glycosylation [9; 11]. PrLZ contained a leucine zipper in the central region and two PEST domains in the N- and C-terminal regions, respectively (Figure 2). Clusters of serine- and threonine-phosphorylation sites were seen upstream from and within the two PEST domains. In addition, three consensus N-glycosylation sites were present in PrLZ (Figure 2). It appeared that PrLZ were subjected to multiple protein modifications.
Many homologous sequences were identified when PrLZ isoforms were used to search the GenBank. Figure 2 lists a few representative sequences to show structural conservation. A structural difference between PrLZ and the prototypic TPD52 was the presence of a unique N-terminus in the PrLZ [9]. This conserved N-terminus could be seen in PrLZ proteins of the mouse, rat, quail, and frog (Figure 2). We carried out RT-PCR cloning and isolated alternatively spliced PrLZ from the rat prostate (GenBank Accession number GQ499323). Both the PrLZ-224 and PrLZ-247 isoforms could be amplified and cloned from rat prostate, suggesting that alternative splicing of PrLZ is a common phenomenon.
The primary structure of PrLZ is highly conserved among species. The amino acid sequence of PrLZ-247, for example, was 87.9% identical between human and rat. When sequence conservation was assessed based on charges [12], more than 90% of the divergences could be categorized as conservative replacements, and sequence similarity between the human and rat rose to 98%. The central region of PrLZ, including the leucine zipper domain had the highest sequence identity among species (Figure 2), suggesting that this region has important biological functions.
4. Interaction of the PrLZ with 14-3-3 proteins
PrLZ contains a central leucine zipper domain known to be an interface for protein-protein interaction [13]. PrLZ may harbor additional sites mediating the interaction with other proteins, since PrLZ contains multiple phosphorylation motifs that may serve as protein-binding sites (Figure 2). In this regard, other members of the TPD52 family have been found to interact with 14-3-3 proteins [14; 15], which bind preferably to sites of phosphrylated serine and threonine [16; 17]. We performed studies to evaluate whether PrLZ had the capability of interacting with 14-3-3 proteins.
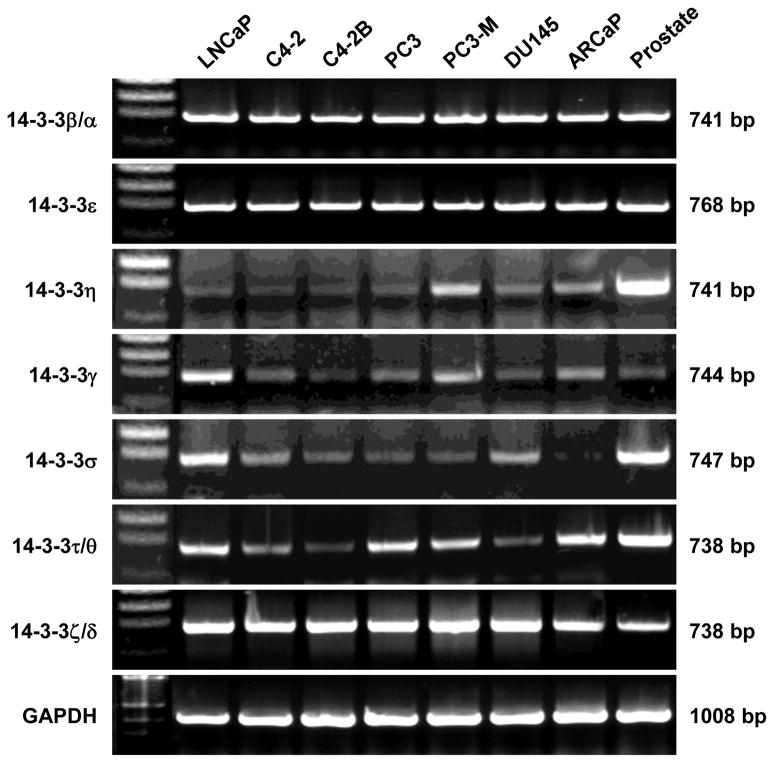
We first examined the expression of 14-3-3 genes in human prostate cancer cell lines by RT-PCR analysis. Total RNA from a normal human prostate was used for comparison. This study revealed that several of the 14-3-3 genes are differentially expressed (Figure 3). Whereas expression of 14-3-3β/α, 14-3-3ε, and 14-3-3ζ/δ appeared ubiquitous, expression of 14-3-3η was decreased in most prostate cancer cell lines compared to normal prostate. Most notably, there was decreased expression of 14-3-3γ, 14-3-3σ, and 14-3-3τ/θ in the LNCaP lineage, where androgen-independent and tumorigenic C4-2 and C4-2B derivative sublines showed lower expression (Figure 3). Reduced 14-3-3 expression correlates with malignant potential in these cells.
Figure 3. Expression of 14-3-3 genes in human prostate cancer cell lines.
Expression of the seven 14-3-3 genes was detected with gene-specific primer pairs in RT-PCR analysis. Commercial total RNA of a normal human prostate (Prostate) was used as control. Representative results from two experiments are shown.
We then used mammalian two-hybrid assays to assess the interaction between PrLZ and 14-3-3 proteins. Coding sequences for the five PrLZ isoforms were cloned individually to the pACT and pBIND vectors as fusion proteins. Coding sequences for the seven 14-3-3 proteins were obtained from C4-2 cells by RT-PCR cloning, and transferred to the pACT and pBIND vectors after DNA sequencing. In mammalian two-hybrid assays in C4-2 cells, each PrLZ isoform in pACT was examined for its interaction with the seven 14-3-3 proteins in pBIND. In a complimentary study, each PrLZ isoform in pBIND was examined for interaction with the seven 14-3-3 proteins in pACT.
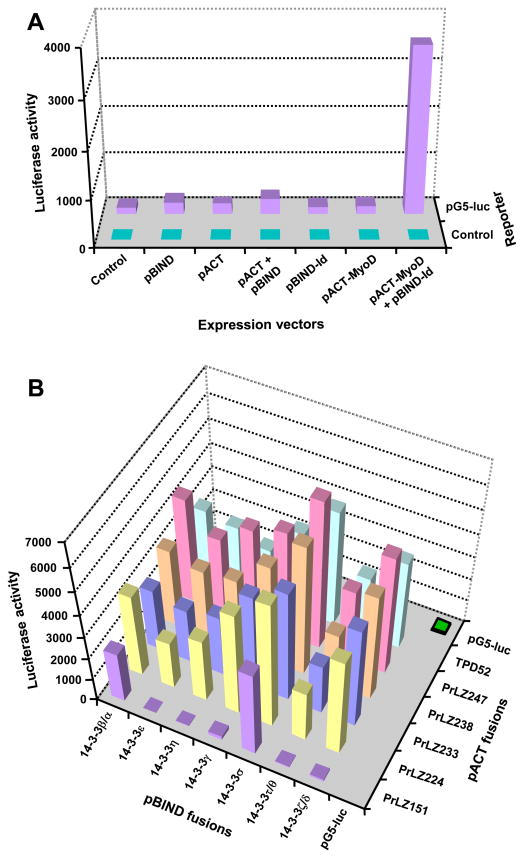
A series of control assays showed that in C4-2 cells, specific protein-protein interaction led to high luciferase activity of the pG5-luc reporter (Figure 4A), while little effect was detected by transfecting empty vectors (Figure 4A) or individual constructs containing PrLZ or 14-3-3 coding sequences (data not shown). On the other hand, specific luciferase activities were detected when a PrLZ fusion construct in pACT was combined with a 14-3-3 fusion construct in pBIND in co-transfection (Figure 4B). Similar results were obtained in the complementary experiments (data not shown).
Figure 4. Interaction between PrLZ and 14-3-3 proteins as detected with mammalian two-hybrid assay.
A, the assay was calibrated with empty vectors of the two-hybrid system and with kit controls (pACT-MyoD and pBIND-Id). B, results of the PrLZ isoforms in pACT co-transfected with each 14-3-3 cDNA in pBIND. TPD52, the prototype of the TPD52 family, was included in the study. Similar results were obtained in a complementary experiment, when PrLZ isoforms in pBIND was examined for interaction with 14-3-3 in pACT (data not shown). When transfected individually, neither the PrLZ nor 14-3-3 constructs induced significant luciferase activity from the pG5-luc reporter (data not shown). Each data represent the mean of triplicate assays. Standard deviation was less than 5% of the mean and is not shown.
Mammalian two-hybrid assays revealed complex interactions between PrLZ isoforms and 14-3-3 proteins. In terms of the PrLZ isoforms, the reporter activity seemed to be correlated to size of the isoform, with the longest PrLZ-247 showing the highest activity in general, and the shorter PrLZ-224 displaying less activity. The shortest PrLZ-151, which lacks the C-terminal region found in other isoforms, showed specific interaction only with 14-3-3β/α and 14-3-3σ proteins. These results suggested that multiple sites in PrLZ may be involved in the interaction with 14-3-3 proteins, and the N- and C-terminal regions of PrLZ could separately mediate the interaction.
Among 14-3-3 proteins, there was a clear difference in terms of the capability to interact with PrLZ isoforms. 14-3-3σ had the highest capability, followed by 14-3-3β/α, 14-3-3ζ/δ, and 14-3-3γ, while 14-3-3τ/θ showed the lowest interaction (Figure 4B). Only 14-3-3β/α and 14-3-3σ could interact with the PrLZ-151 isoform, suggesting that these two proteins harbor unique features for interacting with the N-terminal region of the PrLZ.
Discussion
Our previous studies demonstrated that the prostate-specific and androgen-regulated PrLZ gene is a potential marker for prostate cancer and a candidate oncogene involved in prostate cancer development, progression, and metastasis [5; 9]. In this study, we carried out structural comparisons of the PrLZ gene and its transcripts. Three additional coding exons were identified and five PrLZ isoforms resulting from alternative splicing were recognized (Figure 1A, 1B, and Figure 2). In studies exploring the mode of action, we found that PrLZ isoforms interact with 14-3-3 proteins. Results from this work unveil a complex regulatory mechanism for prostate-specific genes and laid a foundation for elucidating the role of PrLZ in regulating prostate morphogenesis and in promoting prostate cancer development and progression.
Alternative splicing leads to the production of multiple proteins from a single gene. Since the resultant proteins contain altered structure and divergent function, regulated alternative splicing is a critical mechanism for fine tuning gene function during development and for maintaining functional homeostasis [18; 19]. We have reported that during early embryonic prostate morphogenesis, PrLZ protein is localized mainly in the nucleus of epithelial cells, while a high level of PrLZ is seen in both the nuclear and cytoplasmic compartments in young adults [5]. It is mandatory to investigate whether alternative splicing of the PrLZ is regulated developmentally, and whether individual isoforms are produced at specific stages of prostate development and have different subcellular localization.
Abnormal splicing is found to cause genetic diseases [20]. In cancers, abnormal splicing can cause tumor development [21] and tumor cell heterogeneity [22]. With the potential to produce five isoforms, the PrLZ gene is an ideal model for investigating the role of abnormal alternative splicing in prostate cancer development and progression. Although high levels of PrLZ expression are seen during normal prostate development and in prostate cancer, the isoforms expressed in normal tissues may be different from those found in cancer cells. Further studies are warranted to compare the alternative splicing of PrLZ between normal and cancer cells using the RT-PCR detection method shown in Figure 1C. With coding sequences for the five isoforms cloned, we are currently comparing the effect of individual PrLZ isoforms on the proliferation and survival of prostate cancer cells.
The mechanism by which PrLZ promotes cancer cell growth and survival has yet to be elucidated. Although PrLZ proteins contain a leucine zipper and are present in the nucleus, none of the PrLZ isoforms contains a consensus DNA-binding motif and they may not be transcription factors. Instead, both our published works and preliminary results suggest that PrLZ exerts an effect on proliferation and survival by affecting the subcellular localization, availability, and functional activity of other proteins. In this regard, 14-3-3 proteins are known binding partners of the TPD52 family [14; 15]. Accordingly, we initiated an examination of PrLZ interaction with other proteins by exploring the interaction between PrLZ and 14-3-3 proteins.
The 14-3-3 proteins are a family of regulatory proteins that play complex roles in cancer development [16; 17]. We found that three 14-3-3 genes (14-3-3η, 14-3-3σ, and 14-3-3τ/θ) were down-regulated in prostate cancer (Figure 3). Except for 14-3-3ζ/δ, we did not find substantially increased expression of other 14-3-3 genes in the prostate cancer cell lines as compared to normal prostate. In the LNCaP lineage, the trend of the 14-3-3 expression is the opposite of the enhanced PrLZ expression, suggesting an imbalanced level of these proteins in prostate cancers. Interestingly, the loss of 14-3-3σ is a recurrent scenario in malignancies [23; 24].
In a comprehensive assessment of the interaction between PrLZ and 14-3-3 proteins, we detected specific reporter luciferase activities that indicated interactions between these proteins (Figure 4). On the other hand, the significance of the interactions remains unclear. PrLZ and 14-3-3 proteins may have mutually antagonistic functions. Increased levels of PrLZ in prostate cancer may result in sequestration of 14-3-3 proteins, blocking the tumor suppressor function of 14-3-3. On the other hand, it is also possible that both PrLZ and 14-3-3 have tumor promoting activity. Increased levels of PrLZ may compensate for the loss of 14-3-3 expression in cancer cells for cell proliferation and survival. Further investigation is warranted to elucidate the functional significance of the interaction between PrLZ and 14-3-3 proteins.
Supplementary Material
Acknowledgments
This work is supported by esearch grants R21CA112330, PC040578, CA132388 (RXW), and CA98912-02 (LWKC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eder IE, Bektic J, Haag P, Bartsch G, Klocker H. Genes differentially expressed in prostate cancer. BJU Int. 2004;93:1151–5. doi: 10.1111/j.1464-410X.2004.04869.x. [DOI] [PubMed] [Google Scholar]
- 2.Gelmann EP, Semmes OJ. Expression of genes and proteins specific for prostate cancer. J Urol. 2004;172:S23-6. doi: 10.1097/01.ju.0000141781.70474.48. discussion S26-7. [DOI] [PubMed] [Google Scholar]
- 3.Myers RB, Grizzle WE. Changes in biomarker expression in the development of prostatic adenocarcinoma. Biotech Histochem. 1997;72:86–95. doi: 10.3109/10520299709082217. [DOI] [PubMed] [Google Scholar]
- 4.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, Sesterhenn IA, Srikantan V, Moul JW, Srivastava S. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–11. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 5.Wang R, Xu J, Mabjeesh N, Zhu G, Zhou J, Amin M, He D, Marshall FF, Zhau HE, Chung LW. PrLZ is expressed in normal prostate development and in human prostate cancer progression. Clin Cancer Res. 2007;13:6040–8. doi: 10.1158/1078-0432.CCR-07-0640. [DOI] [PubMed] [Google Scholar]
- 6.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–41. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 7.Dachs GU, Dougherty GJ, Stratford IJ, Chaplin DJ. Targeting gene therapy to cancer: a review. Oncol Res. 1997;9:313–25. [PubMed] [Google Scholar]
- 8.Hsieh CL, Chung LW. New prospectives of prostate cancer gene therapy: molecular targets and animal models. Crit Rev Eukaryot Gene Expr. 2001;11:77–120. [PubMed] [Google Scholar]
- 9.Wang R, Xu J, Saramaki O, Visakorpi T, Sutherland WM, Zhou J, Sen B, Lim SD, Mabjeesh N, Amin M, Dong JT, Petros JA, Nelson PS, Marshall FF, Zhau HE, Chung LW. PrLZ, a novel prostate-specific and androgen-responsive gene of the TPD52 family, amplified in chromosome 8q21.1 and overexpressed in human prostate cancer. Cancer Res. 2004;64:1589–94. doi: 10.1158/0008-5472.can-03-3331. [DOI] [PubMed] [Google Scholar]
- 10.Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–7. [PubMed] [Google Scholar]
- 11.Byrne JA, Mattei MG, Basset P. Definition of the tumor protein D52 (TPD52) gene family through cloning of D52 homologues in human (hD53) and mouse (mD52) Genomics. 1996;35:523–32. doi: 10.1006/geno.1996.0393. [DOI] [PubMed] [Google Scholar]
- 12.Dagan T, Talmor Y, Graur D. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol Biol Evol. 2002;19:1022–5. doi: 10.1093/oxfordjournals.molbev.a004161. [DOI] [PubMed] [Google Scholar]
- 13.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–6. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 14.Boutros R, Bailey AM, Wilson SH, Byrne JA. Alternative splicing as a mechanism for regulating 14-3-3 binding: interactions between hD53 (TPD52L1) and 14-3-3 proteins. J Mol Biol. 2003;332:675–87. doi: 10.1016/s0022-2836(03)00944-6. [DOI] [PubMed] [Google Scholar]
- 15.Boutros R, Byrne JA. D53 (TPD52L1) is a cell cycle-regulated protein maximally expressed at the G2-M transition in breast cancer cells. Exp Cell Res. 2005;310:152–65. doi: 10.1016/j.yexcr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Tzivion G, Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–4. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 17.Yaffe MB, Elia AE. Phosphoserine/threonine-binding domains. Curr Opin Cell Biol. 2001;13:131–8. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- 18.Irimia M, Rukov JL, Roy SW, Vinther J, Garcia-Fernandez J. Quantitative regulation of alternative splicing in evolution and development. Bioessays. 2009;31:40–50. doi: 10.1002/bies.080092. [DOI] [PubMed] [Google Scholar]
- 19.Moroy T, Heyd F. The impact of alternative splicing in vivo: mouse models show the way. RNA. 2007;13:1155–71. doi: 10.1261/rna.554607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper TA, Mattox W. The regulation of splice-site selection, and its role in human disease. Am J Hum Genet. 1997;61:259–66. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajan P, Elliott DJ, Robson CN, Leung HY. Alternative splicing and biological heterogeneity in prostate cancer. Nat Rev Urol. 2009;6:454–60. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- 23.Lodygin D, Hermeking H. Epigenetic silencing of 14-3-3sigma in cancer. Semin Cancer Biol. 2006;16:214–24. doi: 10.1016/j.semcancer.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Horie-Inoue K, Inoue S. Epigenetic and proteolytic inactivation of 14-3-3sigma in breast and prostate cancers. Semin Cancer Biol. 2006;16:235–9. doi: 10.1016/j.semcancer.2006.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.