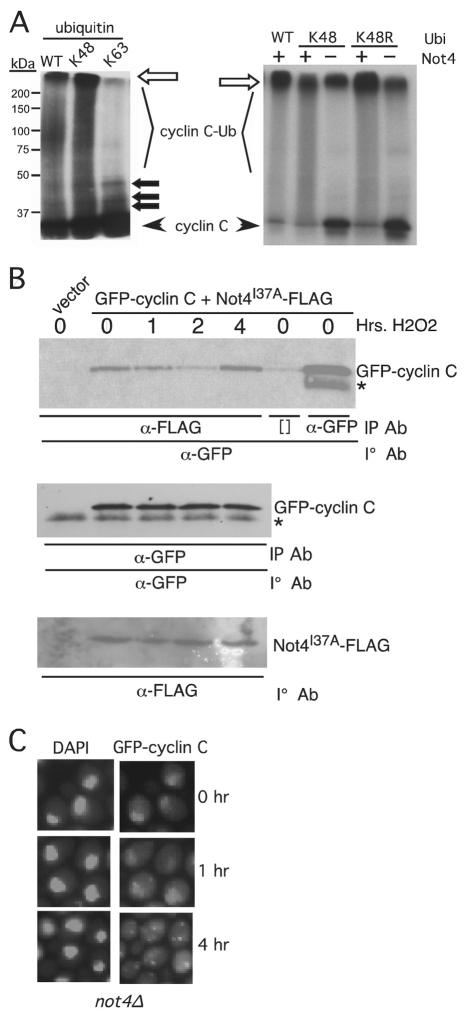
Fig. 7.
Not4p functions in the cytoplasm to promote cyclin C destruction. (A) The in vitro ubiquitylation experiments described in Fig. 2 were repeated with wild-type ubiquitin or derivatives containing only lysine 48 (K48) or lysine 63 (K63) (left panel). The right panel shows ubiquitin in derivatives mutated only for Lys48 (K48R). The unmodified 35S-labeled cyclin C input and ubiquitylated cyclin C species (cyclin C-Ub) are indicated. The open arrow indicates the boundary between the resolving and stacking gel. Closed arrows indicate predicted mono-, bi- and tri-ubiquitylated cyclin C species. The positions of molecular mass markers (kDa) are shown on the left. (B) A wild-type strain (RSY10) expressing GFP–cyclin-C and Not4I37A–FLAG was grown to mid-log-phase and subjected to oxidative stress for the times indicated. Extracts prepared from these samples were immunoprecipitated with anti-FLAG or -GFP monoclonal antibody as indicated and the immunoprecipitates (IP) subjected to western blot analysis probing for GFP–cyclin-C. The input control western blots monitoring GFP–cyclin-C and Not4I37A–FLAG are depicted in the two lower panels. The asterisk indicates a non-specific crossreactive signal. Brackets ([]) indicate the no antibody control lanes. (C) A mutant strain deleted for NOT4 (not4Δ) and expressing GFP–cyclin-C (pBK1) was subjected to H2O2 treatment for the times indicated. The position of the nucleus was determined by DAPI staining.