Abstract
Fluorescence resonance energy transfer (FRET) has been used to study the global folding of an uranyl (UO22+)-specific 39E DNAzyme in the presence of Mg2+, Zn2+, Pb2+, or UO22+. At pH 5.5 and physiological ionic strength (100 mm Na+), two of the three stems in this DNAzyme folded into a compact structure in the presence of Mg2+ or Zn2+. However, no folding occurred in the presence of Pb2+ or UO22+; this is analogous to the “lock-and-key” catalysis mode first observed in the Pb2+-specific 8–17 DNAzyme. However, Mg2+ and Zn2+ exert different effects on the 8–17 and 39E DNAzymes. Whereas Mg2+ or Zn2+-dependent folding promoted 8–17 DNAzyme activity, the 39E DNAzyme folding induced by Mg2+ or Zn2+ inhibited UO22+-specific activity. Group IIA series of metal ions (Mg2+, Ca2+, Sr2+) also caused global folding of the 39E DNAzyme, for which the apparent binding affinity between these metal ions and the DNAzyme decreases as the ionic radius of the metal ions increases. Because the ionic radius of Sr2+ (1.12 Å) is comparable to that of Pb2+ (1.20 Å), but contrary to Pb2+, Sr2+ induces the DNAzyme to fold under identical conditions, ionic size alone cannot account for the unique folding behaviors induced by Pb2+ and UO22+. Under low ionic strength (30 mm Na+), all four metal ions (Mg2+, Zn2+, Pb2+, and UO22+), caused 39E DNAzyme folding, suggesting that metal ions can neutralize the negative charge of DNA-backbone phosphates in addition to playing specific catalytic roles. Mg2+ at low (<2 mm) concentration promoted UO22+-specific activity, whereas Mg2+ at high (>2 mm) concentration inhibited the UO22+-specific activity. Therefore, the lock-and-key mode of DNAzymes depends on ionic strength, and the 39E DNAzyme is in the lock-and-key mode only at ionic strengths of 100 mm or greater.
Keywords: FRET, DNA, DNAzymes, enzymes, metalloenzymes, uranium
Introduction
Metal ions play important roles in biological systems. A more clear understanding of how metal ions bind to biomolecules with high specificity can significantly advance our knowledge of fundamental chemistry and biology, and aid in the development of metal ion sensors, novel biocatalysts, or metal-based pharmaceutical agents.[1] In contrast to the vast amount of information known about metalloproteins, much less is known about how nucleic acids interact with metal ions. Therefore, these interactions are currently being investigated in chemistry and biology fields.[1–2] Catalytic RNA[3] and DNA,[4] discovered in the 1980 s and 1990 s, respectively, are of particular interest because metal ions have been shown to play structural and/or functional roles in these biomolecules.
We are interested in the roles of metal ions in catalytic DNA molecules. These molecules are also called deoxyribozymes and DNAzymes, and will be referred to as DNAzymes herein. Just as with catalytic RNA molecules or ribozymes, DNAzymes have been shown to catalyze many reactions.[1c, 5] However, DNAzymes are much more cost-effective to synthesize and much more stable than catalytic RNA molecules; this makes DNAzymes more suitable for fundamental studies and practical applications. With the further addition of functional groups through chemical modifications, functional DNA molecules with novel functions can be generated by in vitro selection.[6] More importantly, in vitro selection has been used to obtain a number of DNAzymes with high specificity for metal ions, such as Pb2+,[4] Mg2+,[7] Ca2+,[8] Cu2+,[9] Zn2+,[10] Mn2+,[11] UO22+,[12] and Hg2+.[13] These DNAzymes have also been converted into highly sensitive and selective metal-ion sensors[14] and are excellent models for metal-ion interactions with nucleic acids. The insights gained from this study can advance our goal toward the rational design of metal-ion sensors.
The Pb2+-specific 8–17 DNAzyme has been isolated by several different groups through in vitro selection,[7–8, 10–11] and out of all DNAzymes obtained to date, it has been subjected to the most biochemical and biophysical investigations, including metal-dependent structural and functional studies. Biochemical assays have shown that the metal-dependent activities can be ranked as: Pb2+ ≫ Zn2+ ≫ Mn2+ ≈ Co2+ > Ni2+ > Mg2+ ≈ Ca2+ > Sr2+ ≈ Ba2+.[8b, 15] To elucidate the origin of this metal-ion selectivity, the metal-ion-dependent folding of 8–17 DNAzyme has been studied by using fluorescence resonance energy transfer (FRET), both in bulk solution[16] and at the single-molecule level.[17] One interesting finding from these studies is that whereas other, less-active metal ions, such as Mg2+ and Zn2+, induce global folding, no global folding was observed for the most active metal ion–Pb2+. These results point to a possible origin for the high Pb2+-dependent activity: the 8–17 DNAzyme in the resting state forms a structure that binds to Pb2+ selectively, although the DNAzyme is in the resting state, like a lock and key. Recent contact photo-cross-linking studies also found that local instead of global folding is responsible for the Pb2+-dependent activity of the 8–17.[18]
Whereas the “lock-and-key” catalysis mode is common in protein enzymes, this was the first time that it was demonstrated in a DNAzyme system. Most catalytic DNA and RNA molecules assume resting-state conformations that do not support efficient catalysis, and must change conformations to become active.[19] An immediate question is whether the 8–17 DNAzyme is an exception to this rule or if the lock-and-key mode is common among DNAzymes. Furthermore, the Pb2+ concentration used in these studies was much lower (in the micromolar range) than the concentrations of other metal ions (e.g., a millimolar range for Mg2+). Such a low concentration may partially be responsible for the observed difference.
To answer these questions, we turned our attention to a UO22+-specific DNAzyme called 39E, which was isolated by our group through in vitro selection.[12] The sequences of 39E and the substrate 39S are shown in Figure 1 a. The DNA enzyme strand (39E, in green) can form a stem-loop structure with the DNA-substrate strand (39S, in black). The substrate strand is composed of deoxyribonucleotides, except for a single adenosine ribonucleotide in the middle (rA, shown in red). Upon addition of UO22+, 39E catalyzes the phosphodiester transfer reaction at the ribonucleotide scissile site in the 39S; this cleaves the strand into two pieces. This DNAzyme has comparable activity to and even higher metal selectivity than the 8–17 DNAzyme. For example, whereas the 8–17 DNAzyme has some Zn2+- and Mg2+-dependent cleavage activity, the 39E DNAzyme has no detectable activity even in the presence of either of these metal ions in a millimolar concentration range. Among the twenty competing metal ions tested, only Pb2+ caused <10% cleavage at 1 mm.[20] Because of these features, this DNAzyme has been transformed into a fluorescent sensor based on the catalytic beacon method, with a detection limit of 11 ppt (45 pM) and over a millionfold selectivity over competing metal ions.[12] In addition, the 39E DNAzyme has been converted into a colorimetric sensor,[21] a label-free fluorescent sensor,[22] a logic gate,[23] and other sensors.[24] Given the excellent performance and wide range of applications, we wish to investigate whether and how the conformational changes of the 39E–39S complex induced by different metal ions are responsible for the activity and selectivity.
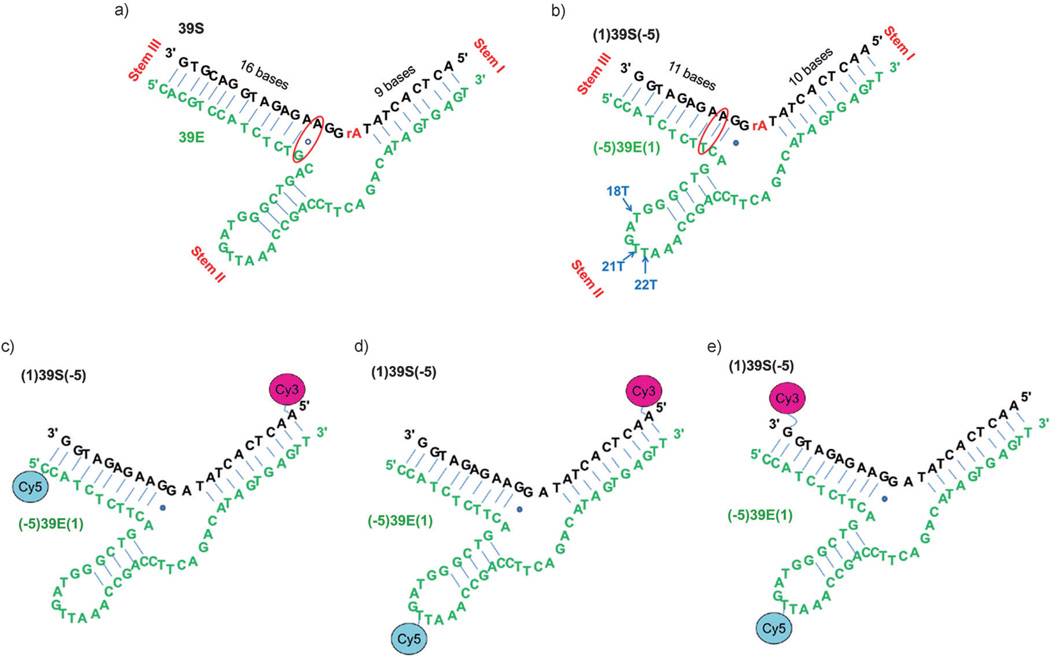
Figure 1.
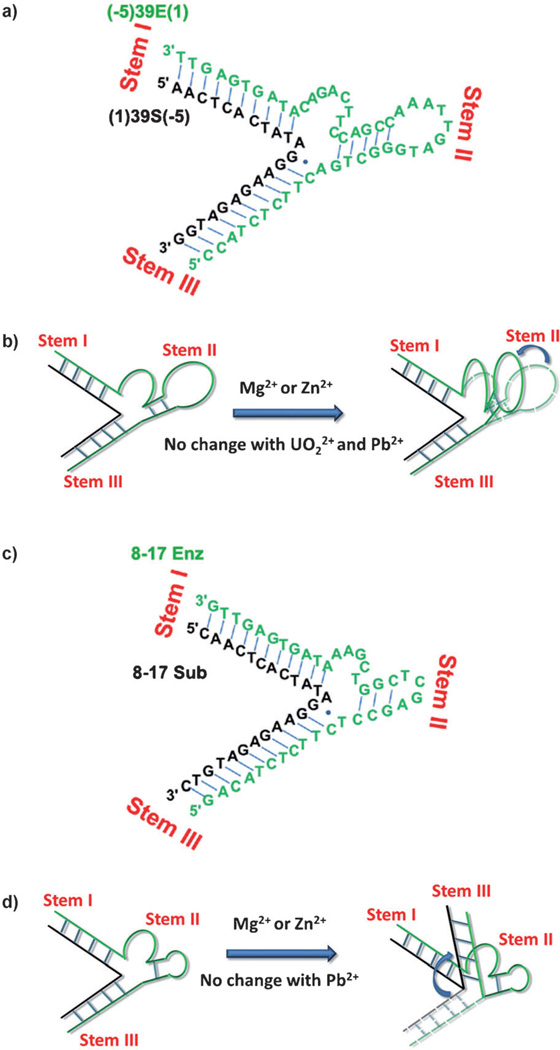
The sequences and secondary structures of the UO22+-specific DNAzyme–substrate complex used previously for fluorescent sensing[12] and the sequences and secondary structures modified for the FRET studies described herein. a) The UO22+-specific DNAzyme–substrate used for fluorescent sensing based on the catalytic beacon method. The substrate strand is shown in black and the enzyme strand is shown in green. The rA base shown in red in the middle of the substrate strand is the cleavage site. b) The modified sequence used in activity assays to identify which position Cy5 could best be attached to for FRET studies. The arms are shortened on the left and extended on the right to make it symmetrical for FRET studies. An A·G wobble pair three bases from the scissile rA is replaced with an A·T pair. The three T bases indicated by arrows are possible positions to introduce Cy5 into the loop (18T, 21T, and 22T). The three arms are termed I (the linear stem composed of the 5′ end of the 39S strand and the 3′ end of the 39E strand), II (the bulged loop in the middle of the 39E strand) and III (the linear stem composed of the 3′ end of the 39S strand and the 5′ end of the 39E strand). c–e) The sequences, secondary structures, and positions of fluorophore pairs for FRET studies involving: c) stems I–III, d) stems I–II, and e) stems II–III. The rA at the cleavage site is replaced by an A in order to probe only conformation changes.
To monitor the DNA or RNA conformational changes induced by metal ions, several methods have been used, including X-ray crystallography,[25] isothermal titration calorimetry,[26] transient electric birefringence,[27] FRET,[16–17, 19b, 28] EPR,[1d, 29] and NMR[30] spectroscopic techniques. We chose FRET because it provides long-range-distance (10–100 Å) information that is helpful in studying the global structure of nucleic acids.[31] FRET monitors the interactions between two fluorophores. When these fluorophores are sufficiently close, the emission from one fluorophore (the donor) excites the other fluorophore (the acceptor).[31] FRET has been widely used to detect the ion-induced folding of the hammerhead ribozyme,[19b, 32] hairpin ribozyme,[33] hepatitis delta virus (HDV) ribozyme,[34] Neurospora Varkud satellite (VS) ribozyme,[35] Group II intron ribozyme,[36] Diels–Alderase ribozyme,[30c, 37] and the DNA and RNA Holliday junctions.[38] FRET has also been used in the study of other RNA or DNA constructs, including the duplex,[39] bulged duplex,[40] triplex,[41] guanine quadruplex,[42] and i-motif.[43] Herein, we report FRET studies of metal-ion-dependent folding of the 39E DNAzyme–substrate complex and correlate this folding with the activity. We found a second example of the lock-and-key binding mode in the 39E DNAzyme in the presence of UO22+, suggesting that the lock-and-key may be a common mode of catalysis, at least for the most efficient DNAzymes. Furthermore, we showed that UO22+ is capable of inducing the 39E–39S complex to fold at low monovalention concentrations, indicating that the DNAzyme–substrate complex can undergo global folding in the presence of UO22+ under low ionic strength when monovalent metal ions are not capable of bringing about the folding. Under conditions of physiological ionic strength (100 mm Na+), however, the 39E–39S complex is fully folded into the most active conformation ready for UO22+ binding.
Results and Discussion
DNAzyme constructs for FRET studies
The 39E construct originally reported was asymmetric, with sixteen base pairs in the left binding arm and nine in the right binding arm around the scissile site (Figure 1 a).[12] This design is important for the catalytic beacon sensor to ensure that both arms of substrate 39S and enzyme 39E are fully hybridized before cleavage, but that the shorter arm dehybridizes after cleavage due to a lower melting temperature. This shorter arm is functionalized with a fluorophore, and when it is released, it produces a fluorescent signal increase.[1c, 14, 44] A recent biochemical study of this DNAzyme indicated that the arm lengths can be varied without affecting the metal-ion-dependent activity.[20] Therefore, to eliminate arm-length-dependent FRET changes that have little to do with metal-ion activity or selectivity, we made the two arms of equal length by shortening the left arm and extending the right arm (Figure 1 b). In addition, the A·G wobble pair three base pairs away from the cleavage site has been found to be nonessential;[20] when this wobble pair was changed to a regular A·T Watson–Crick base pair, the resulting DNAzyme had a similar reactivity to the original construct (the mutant 39E had a kobs of (1.8 ± 0.4) min−1, whereas the original 39E had a kobs of (1.0 ± 0.2) min−1).[20] We thus decided to use the construct with the A·T mutation at this position. Finally, the rA at the cleavage site was replaced with an adenosine deoxyribonucleotide to render the substrate uncleavable so that the FRET studies could focus on metal-ion-dependent folding. The replacement of a ribonucleotide with a deoxyribonucleotide at the scissile position is a common practice in folding studies with FRET.[16b, 19b, 32b, e] Such a practice assumes that replacing the ribonucleotide with a deoxyribonucleotide will not change the overall folding patterns. However, a single-molecule study of the Group II intron ribozyme reported that, by replacing a ribonucleotide cytidine with a deoxyribonucleotide cytidine, the rate of the cleavage reaction was reduced as well as the frequency of high FRET-efficiency (EFRET) populations.[36a] On the other hand, a single-molecule FRET study of the 8–17 DNAzyme with an active ribonucleotide at the scissile position and a bulk FRET study of the same DNAzyme with a deoxyribonucleotide at the same position showed that changing the ribonucleotide did not affect the folding kinetics significantly.[17a]
The FRET construct is shown in Figure 1 b, and stems I, II, and III are labeled according to the naming convention of the hammerhead ribozyme[19b, 32b] and the 8–17 DNAzyme.[16b] To probe the folding of each stem by FRET, the stems were labeled with Cy3 (FRET donor) and Cy5 (FRET acceptor) fluorophores, as shown in Figure 1 c–e. A six-carbon alkyl linker to the fluorophore was used to avoid perturbing the folding of the 39E–39S complex.[45] Of the fluorophore-labeled positions, stem II had the highest probability of interfering with the folding and activity of the DNAzyme–substrate complex, because the fluorophore is in the middle of the DNAzyme strand rather than at the end. To find which position was suitable for internal labeling at stem II, each of the three T bases in the stem II loop (shown in Figure 1 b) was labeled with Cy5 and the activities of the modified enzymes were investigated by using 32P-labeled substrates. All three modified enzymes maintained the activities (kobs are (1.6 ± 0.1), (2.0 ± 0.1), and (1.7 ± 0.1) min−1 at 18T, 21T, and 22T positions, respectively, which is similar to the observed rate of the unmodified construct (kobs is (2.1 ± 0.2) min−1)). Because the 21T modification has the least effect on the activity of the enzyme, it was chosen for all internal labeling.
FRET studies of metal-ion-dependent folding in a buffer containing 100 mm Na+
By labeling the stems two at a time, one with a FRET donor (Cy3) and another with a FRET acceptor (Cy5), the folding of the three stems in the presence of Mg2+, Zn2+, Pb2+, or UO22+ was monitored in a buffer with 100 mm Na+ (50 mm Na-MES buffer, pH 5.5, and 50 mm NaNO3). One example of emission spectra at donor and acceptor excitation wavelengths and the calculation of the EFRET-versus-Mg2+-concentration plot by acceptor normalization ((ratio)A method) under Mg2+-titration conditions are shown in Figure S1 in the Supporting Information.
Folding induced by Mg2+ or Zn2+
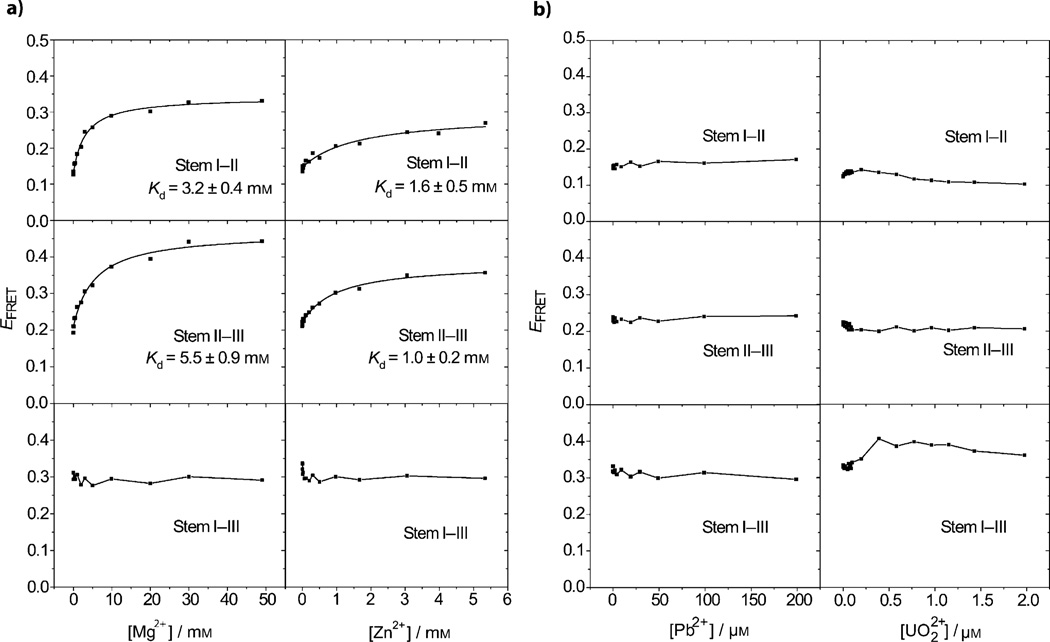
Mg2+ and Zn2+ are common metal cofactors for DNAzymes that play structural or functional roles. For example, Mg2+ induces the folding of a four-way junction in the hairpin[33b] and hammerhead ribozymes.[19b, 32b] The conformation of these and other nucleic-acid enzymes depends on the ionic strength and concentration of specific species (such as surfactants)[46] in the buffer solution. Recent FRET studies of the 8–17 DNAzyme indicated that both Mg2+ and Zn2+ can cause this DNAzyme to fold.[16a,b, 17a] We therefore investigated the Mg2+- and Zn2+-dependent folding of the 39E DNAzyme. As shown in Figure 2 a, the EFRET between stems I–II and II–III increased as the concentration of either Mg2+ or Zn2+ increased, suggesting that these metal ions induced the two stem pairs of the DNAzyme to fold more compactly. On the other hand, the EFRET between stems I–III was largely unaffected by increasing metal-ion concentrations, indicating that this pair did not undergo metal-ion-dependent folding. The apparent dissociation constants were obtained by fitting the folding curves in Figure 2 a, and were generally stronger for Zn2+ ((1.6 ± 0.5) mm between stems I–II and (1.0 ± 0.2) mm between stems II–III) than for Mg2+ ((3.2 ± 0.4) mm between stems I–II and (5.5 ± 0.9) mm between stems II–III). Mg2+ induced a greater change in EFRET; this may correspond to different compact conformations.
Figure 2.
Plots of EFRET versus metal-ion concentrations for different pairs of stems in the buffer containing 100 mm Na+ and a) Mg2+ or Zn2+, or b) Pb2+ or UO22+.
Folding induced by UO22+ or Pb2+
In contrast what was observed for Mg2+- and Zn2+-induced folding, addition of either UO22+ or Pb2+ caused minimal changes in the EFRET within the concentration range that supports the activity of the DNAzyme (Figure 2 b); this suggests that minimal global folding occurred in the presence of these two metal ions.
Because the ionic radii of Mg2+, Zn2+, U (+VI) in UO22+, and Pb2+ are 0.66, 0.74, 0.80, and 1.20 Å, respectively,[47] one explanation for the apparently different folding behaviors of 39E in the presence of these ions is the different sizes or geometries of the metal ions that the DNAzyme interacted with; the greater size of Pb2+ and the different, more linear-shaped UO22+ ions do not induce significant folding of the 39E DNAzyme, whereas the smaller ions Mg2+ and Zn2+ do. To address this question further, we investigated the folding of stem II–III in the presence of the Group IIA series of metal ions in a buffer with 100 mm Na+. The ionic radii for Mg2+, Ca2+, and Sr2+ are 0.66, 0.99, and 1.12 Å, respectively.[47] As shown in Figure S2 in the Supporting Information, all three metal ions promoted folding of 39E. The apparent binding affinity between these metal ions and 39E decreases as the ionic radius of the metal ions increases. Contrary to Pb2+, Sr2+ induces the DNAzyme to fold under the present conditions, in spite of a comparable ionic radius (1.12 Å) to Pb2+ (1.20 Å); hence, ion size alone cannot account for the unique folding behaviors of Pb2+ and UO22+ in the 39E DNAzyme.
Correlation of folding with activity in a buffer containing 100 mm Na+
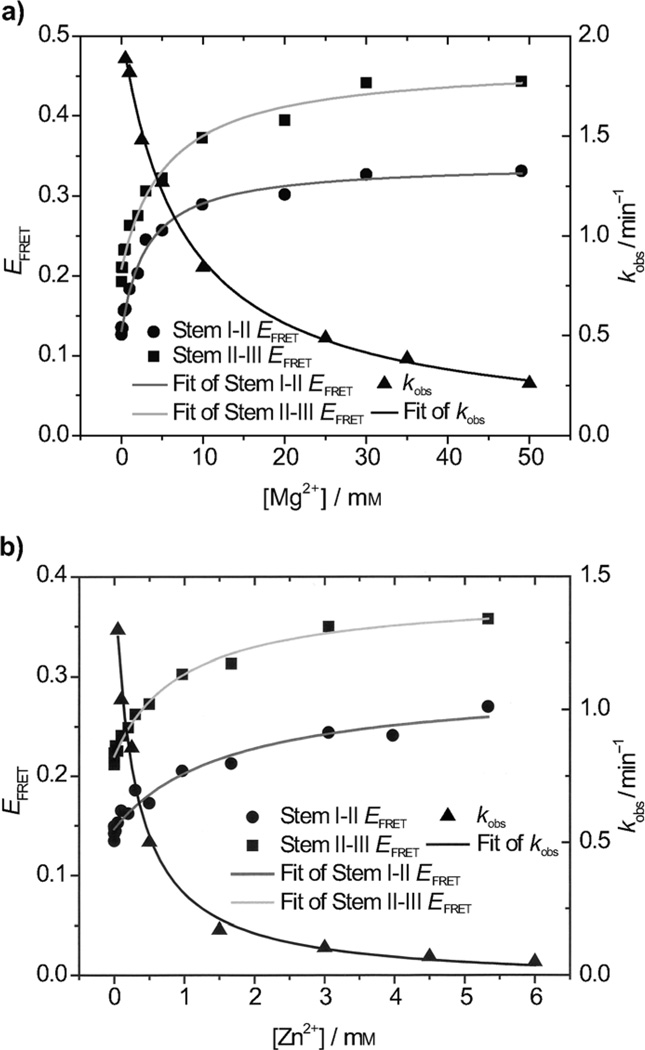
With the metal-ion-induced folding results in hand, we moved on to consider structural information of 39E in the context of the enzymatic activity results.[48] The 39E DNAzyme is highly specific for UO22+ and has no observable activity in the presence of Mg2+ or Zn2+. For example, whereas 90% of the substrate was cleaved by 4 µm UO22+ in 10 min, no cleavage was observed over the course of two hours in the presence of either 50 mm Mg2+ or 6 mm Zn2+ (i.e., the highest concentrations of these two metal ions used in the FRET experiments). As a result, it is difficult to correlate metal-ion-dependent folding with cleavage activity directly. Nonetheless, the correlation is still possible in competition experiments. As demonstrated above, even though Mg2+ and Zn2+ induced stems I–II and II–III of the 39E–39S complex to fold, the enzyme was not active. We then wondered if Mg2+ and Zn2+ in the same concentration range would affect the UO22+-dependent activity of 39E. To answer this question, we carried out activity assays in the presence of 4 µm UO22+ and various concentrations of Mg2+ or Zn2+. Interestingly, the UO22+-dependent activity of 39E decreased as the Mg2+ or Zn2+ concentrations increased (Figure 3), suggesting an inverse correlation between the folding ability and activity of this DNAzyme. More importantly, the dissociation constants (Kd, refer to Table 1) obtained from these inhibition reactions correlated with those obtained from FRET studies. For example, the Kd for Mg2+ inhibition is (7.5 ± 0.8) mm, whereas the Kd for the Mg2+-induced folding of stems I–II and II–III are (3.2 ± 0.4) mm and (5.5 ± 0.9) mm, respectively. On the other hand, the Kd for Zn2+ inhibition is (0.28 ± 0.06) mm, whereas the Kd for the Zn2+-induced folding of stems I–II and II–III are (1.6 ± 0.5) mm, and (1.0 ± 0.2) mm, respectively. These results suggest that the Mg2+- and Zn2+-induced folding of 39E inhibits the UO22+-dependent activity. The Kd values from Zn2+ folding and inhibition experiments are all smaller than those for Mg2+. Many properties of metal ions will affect metalion and nucleic-acid recognition efficiencies, such as charge, size, and coordination number.[2d] These two metal ions have the same ionic charge (+2) and similar ionic radii (0.66 Å for Mg2+ and 0.74 Å for Zn2+), but Zn2+ has an intrinsically higher affinity for ligands than Mg2+. For example, Zn2+ has a stronger affinity for several binding sites of individual nucleic- acid bases.[1j] In this context, these results also suggest that Zn2+ binds more specifically to 39E than Mg2+ does; this is similar to what was found in the 8–17 DNAzyme study.[16b]
Figure 3.
Correlation of the EFRET of the two folding stems and UO22+-induced cleavage rates in the presence of different concentrations of other fold-inducing metal ions (Mg2+ and Zn2+) in a buffer with 100 mm Na+. The black line shows the kobs changes according to the fold-inducing-metal-ion (Mg2+ or Zn2+) concentrations. The grey and light grey curves show the EFRET fittings of stems I–II and II–III versus the fold-inducing metal-ion concentrations. Cleavable substrates with a rA base at the cleavage site was used and the UO22+ concentration in the assays was 4 µm.
Table 1.
A summary of the Kd values from fittings of Mg2+ or Zn2+-dependent FRET curves of different stem pairs and the Mg2+ or Zn2+-dependent inhibition of UO22+-catalyzed activity in the presence of a buffer with 100 mm Na+.
|
Kd [mm] (Stem I–II EFRET) |
Kd [mm] (Stem II–III EFRET) |
Kd [mm] (Stem I–III EFRET) |
Kd [mm] (Activity)[a] |
|
|---|---|---|---|---|
| Mg2+ | 3.2 ± 0.4 | 5.5 ± 0.9 | no folding | 7.5 ± 0.8 |
| Zn2+ | 1.6 ± 0.5 | 1.0 ± 0.2 | no folding | 0.28 ± 0.06 |
This column lists Kd values based on the fitting of the Mg2+ or Zn2+ inhibition to UO22+ cleavage kobs curves.
FRET studies of metal-ion-dependent folding in a buffer containing 30 mm Na+
Although it is interesting to find a second example of the lock-and-key mode of catalysis in a DNAzyme, there is one qualifier, that is, the most active metal ions do not induce folding, but do induce enzyme activity (UO22+ and Pb2+). However, they were investigated at much lower concentrations than the metal ions that induce folding (Mg2+ and Zn2+); the former metal ions were used in nano- to micromolar ranges, whereas the latter were used in the millimolar range. These concentration ranges were chosen because they are the concentration ranges at which the respective DNAzyme activities are induced. Such a difference in concentrations does raise the question of whether and how electrostatic interactions play a role in the folding and activity of DNAzymes. To answer these questions, we turned our attention to the ionic strength at which the DNAzyme foldings and activities were carried out. The FRET studies of the 39E DNAzyme system reported above was performed in a buffer with 100 mm Na+ (50 mm NaNO3 and 50 mm Na-MES, pH 5.5). The conformations of hammerhead ribozyme mutants with three arms selectively elongated were studied by gel electrophoresis under three conditions: in the absence of added ions, with Mg2+, and with Na+. These gel-electrophoresis studies indicated that the global folding of the hammerhead ribozyme was highly dependent on the concentration and identity of the metal ions in the environment.[49] It was found that Na+ alone was not sufficient to induce the compact folding conformation that Mg2+ did. On the other hand, the presence of Na+ did not prevent the Mg2+-induced folding process from occurring. Also, the Holliday junction appeared to assume different conformations at high and low salt concentrations.[38c] In addition, FRET studies with RNase P RNA revealed that the concentration of Mg2+ critically affected the early conformation evolution.[50] Recent experimental and simulation studies have demonstrated that certain monovalent metal ions are also capable of inducing similar folding events and even cleavage activity in many ribozymes[33b, 51] and in the 8–17 DNAzyme.[52] Therefore, we hypothesize the reason that the stem I–III did not fold after the addition of Mg2+ or Zn2+ and that no folding was observed in any of the stems after adding UO22+ and Pb2+, in due to the fact that the DNAzyme had already folded into a defined conformation in the presence of the high concentration of monovalent ions (100 mm Na+) in the buffer. Evidence that supports this hypothesis is the high initial EFRET between stems I–III (ca. 0.3). To test this hypothesis, we lowered the concentration of Na+ in the buffer to 30 mm by using 30 mm Na-MES, pH 5.5, and repeated the FRET studies and activity assays. The concentration of Na+ was not zero because some ionic strength is needed to maintain DNA hybridization.
Mg2+- and Zn2+-induced folding in a buffer containing 30 mm Na+
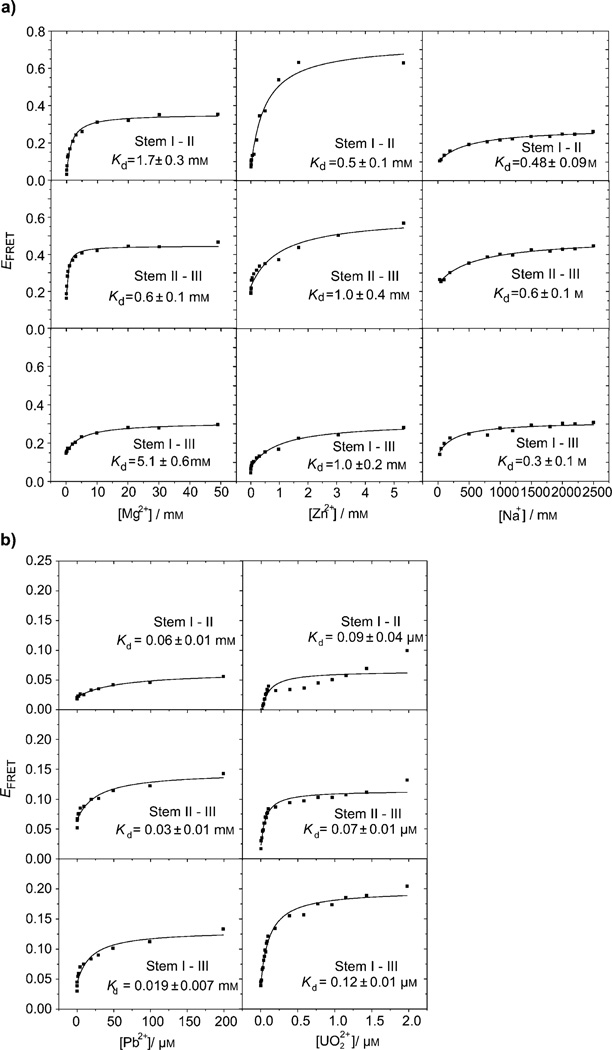
As shown in Figure 2 a, in a buffer with 100 mm Na+, foldings were observed between two sets of arms, except stems I–III. In contrast, when the same experiment was carried out in the presence of a buffer with 30 mm Na+, the EFRET between all three pairs of stems, including stems I–III, were observed to increase as the concentration of Mg2+ or Zn2+ increased (see Figure 4 a). Whereas the folding of stems I–II and stems II–III were already obvious in the buffer with 100 mm Na+, the metal ions bound 39E tighter in the buffer with 30 mm Na+ (see the dissociation constants (Kd) listed in Tables 2 and 3). These results suggest that the ionic strength provided by monovalent ions, such as Na+ also affects the folding of 39E.
Figure 4.
Plots of EFRET versus metal-ion (Mg2+, Zn2+, Na+, UO22+, and Pb2+) concentrations for different stems in a buffer with 30 mm Na+ and a) Mg2+, Zn2+, and Na+, or b)UO22+ and Pb2+.
Table 2.
A comparison of the Kd values obtained by fitting Mg2+-dependent FRET curves of different pairs of stems and the Mg2+-dependent inhibition of UO22+-induced catalytic activity in the presence of a buffer with either 100 mm or 30 mm Na+.
| Mg2+ Titration | Stem I–II | Stem II–III | Stem I–III | Activity[a] |
|---|---|---|---|---|
| Kd [mm] (100 mm Na+) | 3.2 ± 0.4 | 5.5 ± 0.9 | no folding | 7.5 ± 0.8 |
| Kd [mm] (30 mm Na+) | 1.7 ± 0.3 | 0.6 ± 0.1 | 5.1 ± 0.6 | 3.9 ± 0.7 |
This column lists Kd values based on the fitting of the Mg2+ inhibition to UO22+ cleavage kobs curves. In the case of the buffer with 30 mm Na+, only the region in which kobs is decreasing is used for the fitting.
Table 3.
A comparison of the binding affinities of the stems when titrating Zn2+ in a buffer with 100 mm or 30 mm Na+.
| Zn2+ Titration | Stem I–II | Stem II–III | Stem I–III |
|---|---|---|---|
| Kd [mm] (100 mm Na+) | 1.6 ± 0.5 | 1.0 ± 0.2 | no folding |
| Kd [mm] (30 mm Na+) | 0.5 ± 0.1 | 1.0 ± 0.4 | 1.0 ± 0.2 |
In the general polyelectrolyte theory, counter-ions provided by, for example, metal-ion binding or condensation onto DNA, has been classified into two categories: site-bounded (inner sphere) and territorially bounded (outer sphere).[2a] Metal ions that participate in nucleic-acid enzymatic catalysis can also be classified to have catalytic or structural roles.[2d, 53] Because of the phosphate groups, the backbone of DNA is negatively charged. Therefore, a certain number of positively charged ions are needed for two DNA strands to overcome the electrostatic repulsion and form a complex. Monovalent ions present in the buffer normally play a significant structural role in DNA,[33b] although they sometimes play a catalytic role in DNAzymes, as some DNAzymes have been selected in the absence of divalent metal ions.[54] Divalent ions normally play catalytic roles, but they can also bind DNA nonspecifically to serve in a structural role. Because of charge differences and other related properties, these two types of ions may bind DNA at different locations.[55] In a single-molecule FRET study on a two-way junction hairpin ribozyme, monovalent and divalent ions were found to compete to interact nonspecifically with RNA.[56]
Pb2+- and UO22+-induced folding in a buffer containing 30 mm Na+
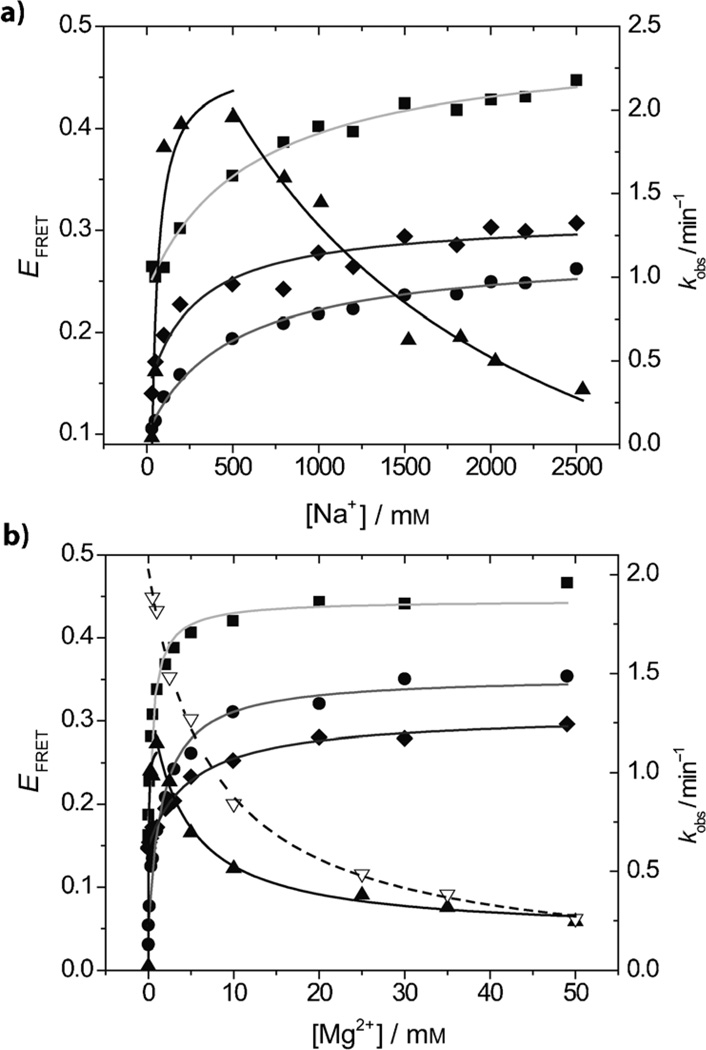
In a buffer with 100 mm Na+, no Pb2+- or UO22+-dependent global folding was observed by FRET (Figure 2 b). In the buffer with 30 mm Na+, however, both Pb2+- and UO22+-dependent global foldings were observed (Figure 4 b). All three pairs of stems became more compact as the concentrations of Pb2+ or UO22+ increased. The apparent Kd values for the global foldings of stems I–II, II–III, and I–III are (0.06 ± 0.01), (0.03 ± 0.01), and (0.019 ± 0.007) mm, respectively for Pb2+, and (0.09 ± 0.04), (0.07 ± 0.01), and (0.12 ± 0.01) µm, respectively, for UO22+. These results strongly suggest that Pb2+ and UO22+ are capable of inducing global folding at very low concentrations if there are not enough other metal ions to fulfill the structural role. Importantly, Pb2+- and UO22+-induced folding is shown to be less significant than the folding induced by other metal ions (e.g., Mg2+ and Zn2+) because the EFRET change in the case of Pb2+ and UO22+ is around 0.1 and resembles the Na+-induced folding. Therefore the lock-and-key catalysis mode depends on the conditions under which the investigations were carried out. The 39E operates in a lock-and-key catalysis mode for UO22+ when a buffer with 100 mm Na+ is present (50 mm NaNO3 and 50 mm Na-MES, pH 5.5). Interestingly, even though Pb2+ induces global folding in the 39E–substrate complex as UO22+ does (i.e., there is no observed folding in the buffer containing 100 mm Na+ and some folding in 30 mm Na+), it does not confer any significant cleavage activity. The difference between the roles played by Pb2+ and UO22+ must be due to a specific binding site; Pb2+ must not occupy the same binding site as UO22+ does in the case of the 39E DNAzyme.
FRET studies of monovalent-metal-ion-dependent folding
Because the previous experiment suggested that different concentrations of Na+ affect the 39E–39S folding differently, we carried out systematic FRET studies of Na+-dependent folding of the three stems. As shown in Figure 4 a, the EFRET between all stems (I–II, II–III, and I–III), increases as the concentration of Na+ increases, with similar binding affinities (Kd(I–II) = (0.48 ± 0.09) m, Kd(II–III) = 0.6 ± 0.1 m, Kd-(I–III) = 0.3 ± 0.1 m). Comparing the initial EFRET values in Figure 2 (0.15, 0.2, and 0.3 for stems I–II, II–III, and I–III) and saturated EFRET values (Na+ titration) in Figure 4 a (0.3, 0.4, and 0.3 for stems I–II, II–III and I–III) indicates that a buffer with 100 mm Na+ is sufficient to fold stem I–III and partially fold stems I–II and II–III.
Correlating the folding of 39E–39S with UO22+ activity in a buffer containing 30 mm Na+
The UO22+-dependent enzyme cleavage activities of 39E were measured under different Na+ concentrations ranging from 30 to 2500 mm (Figure 5 a; 4 µm UO22+ was used). The upper limit was 2500 mm, which is the highest Na+ concentration that has been used in folding studies (Figure 4 a). In the buffer with 30 mm Na+, the initial kobs of the 39E DNAzyme was very small, about 0.04. As the Na+ concentration increased, the kobs rapidly increased, then plateaued between 100 and 500 mm, after which it decreased. This trend revealed a correlation between the Na+-dependent folding and 39E UO22+-dependent cleavage activity. At the beginning, 30 mm Na+ was not enough to neutralize the backbone of the 39E–39S complex and hence did not allow the active conformation to be formed; this resulted in low activity. As the Na+ concentration increased, more and more positive ions were provided and the complex gradually folded into the correct conformation; the enzyme activity recovered accordingly. After reaching the optimal concentration, additional Na+ probably induced alternative folding that interfered with the activity of the 39E DNAzyme.
Figure 5.
The correlation between the EFRET of the three folding stems of 39E and the UO22+-induced cleavage rates in the presence of different concentrations of Na+ and Mg2+ and a 30 mm background Na+ concentration. a) Na+ titration: The black line shows the kobs (▲) changes according to the fold-inducing-metal-ion (Na+) concentration. The grey, light grey, and dark grey curves show the fittings of EFRET between stems I–II (●), II–III (■), and I–III (♦) versus the fold-inducing-metal-ion (Na+) concentrations. b) Mg2+ titration: The black solid line and dashed line show the kobs changes according to the fold-inducing-metal-ion (Mg2+) concentration under buffers containing 30 (▲) and 100 mm Na+ (▽), respectively. The grey, light grey, and dark grey curves show the fittings of EFRET between stems I–II (●), II–III (■), and I–III (♦) versus the fold-inducing-metal-ion (Mg2+) concentrations. The cleavable substrate with a rA base at the cleavage site was used and the UO22+ concentration used in these assays was 4 µm.
Correlation of 39E folding with the UO22+-dependent activity in the presence of different concentrations of Mg2+ and 30 mm Na+
Similar to the trend in the Na+-dependent activity assay, the kobs of the UO22+-specific cleavage in the presence of the buffer with 30 mm Na+ increased rapidly with the Mg2+ concentration until the Mg2+ concentration reached approximately 2 mm, after which kobs decreased (Figure 5 b). To the best of our knowledge, this is the first time that Mg2+ is observed to promote catalysis at low concentrations, but to inhibit catalytic reactions at high concentrations. As explained above in the Na+ study, the DNAzyme proceeded through three stages as Mg2+ was added: a less-active state (the DNAzyme was incompletely folded), an active state (the DNAzyme was sufficient folded when there was ca. 2 mm Mg2+), and a less active state (the DNAzyme was folded unproductively). In contrast to the gradual transitions seen in the Na+ study, the transitions in the Mg2+ case was rather rapid; instead of reaching a plateau at 2 mm, the kobs decreased dramatically. This contrast suggests that Na+ does not neutralize the charge of the nucleic acids as well as Mg2+ does. These results agree with results previously obtained with several RNA structures. Because Mg2+ is divalent, it binds to folded RNA more readily than Na+ does.[2d] Another interesting observation is that the kobs values of 39E at each Mg2+ concentration are lower than the kobs values measured in the buffer with 100 mm Na+. This result makes sense, because, from the FRET data, the binding affinities of 39E for Mg2+ in a buffer with 30 mm Na+ are much greater than the affinities for Mg2+ in a buffer with 100 mm Na+. In Figure 5 b, the kobs results measured in buffers with 100 and 30 mm Na+ are also fitted for comparison. The Kd values obtained by fitting kobs measured in buffers with both 30 and 100 mm Na+ (Table 2) also support the conclusion that the 39E DNAzyme adopts a conformation that binds more tightly to Mg2+ in a buffer with 30 mm Na+ than in a buffer with 100 mm Na+. Interestingly, despite the difference in kobs between the buffers with 30 and 100 mm Na+ at low Mg2+ concentrations, the kobs fitting curves converge at the highest concentration (50 mm Mg2+), suggesting that the initial small ionic strength difference provided by insufficient monovalent ions is compensated by the increasing concentration of Mg2+, and is eventually dominated by the high concentration of divalent metal ions. These different activity trends provide further evidence for the transition from diffusive-binding to site-binding modes.
Comparison between the 39E and 8–17 DNAzymes
The UO22+-specific 39E DNAzyme and the Pb2+-specific 8–17 DNAzyme are two efficient metal-ion-specific DNAzymes with similar secondary structures. Useful insights were obtained into the activity of these enzymes by comparing the similarities and differences in the metal-ion-induced folding patterns at optimal ionic strengths measured by FRET studies:
Mg2+ and Zn2+ induce folding: Mg2+ and Zn2+ can both induce the 39E and 8–17 DNAzymes to fold. The apparent Kd values obtained for 39E were generally lower with Zn2+ ((1.6 ± 0.5) mm between stems I–II and (1.0 ± 0.2) mm between stems II–III) than with Mg2+ ((3.2 ± 0.4) mm between stems I–II and (5.5 ± 0.9) mm between stems II–III); the latter were similar to the Kd values observed for the 8–17 DNAzyme. The difference between the effects of Mg2+ and Zn2+ on the 39E DNAzyme was less pronounced compared with the differences measured for 8–17 DNAzyme (e.g., the Kd for Mg2+ was (1.36 ± 0.24) mm and (0.840 ± 0.023) mm for stem I–III and II–III, respectively, whereas Kd for Zn2+ was (52.6 ± 2.3) µm and (83.2 ± 12.6) µm for stem I–III and II–III, respectively, for 8–17 DNAzyme). In terms of cleavage activity, the 8–17 DNAzyme also displayed a larger difference in Zn2+- and Mg2+-induced cleavage activity compared with 39E DNAzyme, as Zn2+ induces a much faster cleavage rate than Mg2+ does (kobs(2 mm Zn2+) = 2.7 min−1, kobs(2 mm Mg2+) = 0.018 min−1). On the other hand, the 39E DNAzyme appeared to have a much higher selectivity for UO22+ over other metal ions and so the difference between the concentrations needed for Mg2+- and Zn2+-induced folding for FRET results was smaller.
The most active metal ions induce no conformational change: Previous FRET folding studies of the 8–17 DNAzyme indicated that Mg2+ and Zn2+ can cause substantial folding, whereas Pb2+ does not,[16b, 17a] supporting the hypothesis that the 8–17 DNAzyme in the resting state is optimal to support Pb2+-dependent catalysis without the need for global folding. Similarly, the 39E DNAzyme does not show significant conformational changes in the presence of the species that induces the highest activity—UO22+. Because the lock-and-key mode of catalysis has only been demonstrated in one DNAzyme to date, the question remains as to how general this mode of catalysis is in DNAzymes. We have now found a second example of this mode of catalysis; this suggests that it is commonly used by DNAzymes. This conclusion is at least true for some of the most efficient DNAzymes, because both the 8–17 and the 39E DNAzymes fall into this class.
The overall folding schemes for 39E and 8–17 DNAzymes are different: Based on the aforementioned two points, a metal-ion-dependent folding pattern for 39E DNAzyme at optimal ionic strength emerges (Figure 6 b; the 39E DNAzyme sequence is shown in Figure 6 a). For comparison, the folding Scheme of the 8–17 DNAzyme reported previously[16b] is also shown in Figure 6 d (the 8–17 DNAzyme sequence is shown in Figure 6 c). If we use the same nomenclature for the arms of stem I, II, and III, then in response to Mg2+ and Zn2+, stem III of the 8–17 DNAzyme complex folds toward the plane defined by stems I and II. In the 39E DNAzyme, however, in response to Mg2+ and Zn2+, stem II (with the internal loop region) folds towards the plane defined by stems I and III. It is proposed that metal ions could bind to regions of highly negative electrostatic potentials in RNA structures.[2d] These FRET results suggest that each DNAzyme has its own conformation in which a different electrostatic potential exists between different stems. The same metal ions will thus selectively bind to these regions to induce different folding patterns.
The effects of Mg2+- and Zn2+-induced folding on cleavage activity are different: In the case of the 8–17 DNAzyme, Mg2+- and Zn2+-induced folding is conducive for the enzymatic activity, although this activity follows a different reaction pathway than the pathway induced by Pb2+.[16b, 17a] Mg2+ and Zn2+ induce the 8–17 DNAzyme to fold and then induce the cleavage, and the folding and activity were found to be highly correlated; the higher the binding affinity of the FRET-based folding, the higher the binding affinity based on activity.[16b] In the case of the 39E DNAzyme, Mg2+- and Zn2+-induced folding is not conducive for the enzymatic activity of 39E. Mg2+ and Zn2+ can only induce 39E to fold, but cannot lead to the cleavage reaction. More interestingly, Mg2+- and Zn2+-induced folding is counterproductive; this prevents UO22+-dependent cleavage from occurring.
Figure 6.
The folding schemes of the UO22+-specific DNAzyme 39E and the Pb2+-specific DNAzyme 8–17 in response to different metal ions. a) The 39E–substrate sequence used in these FRET experiments; b) the folding scheme of the UO22+-specific DNAzyme 39E–substrate; c) the sequence of the 8–17–substrate sequence used in these FRET experiments; d) the folding scheme of the Pb2+-specific DNAzyme 8–17–substrate. The enzyme strand is shown in green and the substrate strand is shown in black.
Conclusion
In summary, the metal-ion-induced folding of the UO22+-specific DNAzyme 39E has been investigated by FRET and has been correlated with the UO22+-specific cleavage activity in the presence of other metal ions. This study found that the UO22+-specific DNAzyme operates in a lock-and-key mode of catalysis, just as the Pb2+-specific 8–17 DNAzyme does. In contrast to the effects on the 8–17 DNAzyme, however, Mg2+ and Zn2+ induce the 39E–39S complex to assume a conformation that inhibits UO22+-specific activity. By lowering the ionic strength, we have separated the roles of metal ions (such as Na+) that act as nonspecific electrostatic neutralizers from those (such as UO22+) that are selective metal cofactors for enzymatic activities; other metal ions (such as Mg2+) are shown to go through a transition from general charge neutralizers to more specific binders. Therefore, whether or not a DNAzyme will operate in a lock-and-key catalytic mode depends on the ionic strength of the environment.
Experimental Section
Materials
All oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA, USA) in a HPLC-purified form. Uranium acetate dihydrate was purchased from Fisher Scientific (Pittsburgh, PA, USA). All other chemicals were purchased from Sigma–Aldrich (St. Louis, MO, USA) and used without further purification.
Activity assays
To determine the effects of the stem arm length on the DNAzyme activity, cleavage activity assays with a cleavable substrate containing a rA base at the cleavage site were carried out under single-turnover conditions by using a 4 µm UO22+ solution. The samples for this assay were prepared at twice the final concentration and were mixed at a 1:1 ratio at the beginning of the assay. To anneal the enzyme–substrate complex before the reaction, 2 nm (2X) of 5′-32P-labeled (1)39S (−5) and 4 µm (2X) of (−5)39E(1) (a shortened version of the original 39E, see Figure.1b) was mixed together in 50 mm sodium 2-(N-morpholino)ethane sulfonic acid (Na-MES, pH 5.5) and 50 mm NaNO3 buffer, put into a boiling water bath for 1 min, and cooled down to room temperature for 1 h. The above annealed DNAzyme–substrate complex was then mixed with 8 µm (2X) of UO22+ solution in a 1: 1 volume ratio to start the reaction. At different time points, aliquots of the reaction solution were taken out and placed into a stop solution containing 8 m urea, 50 mm EDTA, 0.05% xylene cyanol, and 0.05% bromophenol blue. The cleaved and non-cleaved substrates were separated by electrophoresis by using a 20% polyacrylamide denaturing gel, and the band intensities were quantified with a phosphorimager (Storm 840, GE healthcare Life Science).
To correlate the metal-ion-dependent folding from FRET studies with metal-specific activity, a cyanine-fluorophore-labeled DNA substrate was used under similar conditions as in the 32P assay presented above, except 2 µm (2X) of 5′-labeled Cy3-(1)39S(−5) and 3 µm (2X) of 5′-labeled Cy5-(−5)39E(1) were used. Again the assay employs a cleavable substrate with a rA base at the cleavage site. To avoid any interference from organic dyes in the stop solution when measuring the fluorescent signal of the Cy3/Cy5 fluorophore labels on the DNA, a colorless stop solution containing 8 m urea and 50 mm EDTA, without the two organic dyes (xylene cyanol and bromophenol blue), was used. The band intensities were quantified with a phosphorimager (FLA-3000, Fujifilm). In both the 32P- and fluorophore-labeled assays, the fraction of cleaved DNA substrate was plotted against time by using the program SigmaPlot 8.0 (Systat Software) and the curve was fitted with nonlinear regression by using the following equation:
in which y is the percentage of cleaved substrate at time t, y0 is the percentage of cleaved substrate at time t = 0, a is the cleavage percentage achieved at t = ∞, and k is the observed rate constant, kobs.
Effects of Mg2+, Zn2+, and Na+ on UO22+-dependent cleavage activity
The activity assays were performed under conditions similar to those described above, except that various concentrations of Mg2+, Zn2+, or Na+ were added to the DNAzyme samples and this mixture was incubated for 30 min to 1 h to ensure that the DNAzyme had folded before an equal volume of UO22+ solution was added to start the reaction.
FRET experiments
Noncleavable substrate in which the rA at the cleavage site was replaced by an adenosine deoxyribonucleotide (A) was used to probe only the conformation changes. To obtain a homogeneous enzyme substrate complex, 10 µm of Cy5-labeled (−5)39E(1) and 10 µm of Cy3-labeled (1)39S(−5) were annealed in 50 mm Tris/acetic acid (pH 7.5) and 100 mm NaNO3 buffer (100 mm instead of 50 mm NaNO3 was used to increase hybridization efficiency and maintain the Na+ concentration consistency) or 30 mm NaNO3 buffer. The mixture was placed in a boiling water bath for 1 min and cooled down to room temperature over the course of 2 h. The DNA complex solution was then placed at 4°C for 30 min to anneal further. The hybridized DNAzyme–substrate complex was then separated from the unhybridized strands by running it through a 16% polyacrylamide native gel (50 mm Tris/acetic acid buffer, pH 8.0 and 100 mm NaNO3) with a power of 4 W for 10 h at 4°C. After verifying the separation by using a phosphorimager (FLA-3000, Fujifilm), the completely hybridized DNAzyme–substrate-complex band was cut out of the gel, crushed, and soaked by shaking for 2 to 3 h in soaking buffers with either 100 mm Na+ (50 mm Na-MES buffer, pH 5.5, and 50 mm NaNO3) or 30 mm Na+ (30 mm Na-MES, pH 5.5). The DNA samples were then recovered from the supernatant after centrifugation. For each FRET experiment, this native-gel-purified DNAzyme–substrate-complex concentration was decreased to approximately 100 nm with the same soaking buffer. Upon addition of concentrated metal-ion solutions, the fluorescence emission spectroscopy for both Cy3 and Cy5 was collected at each metal-ion concentration with polarization of 54.7° for the emission spectrum. The excitation wavelengths for Cy3 and Cy5 were 513 and 648 nm respectively (see Figure S1 in the Supporting Information for donor and acceptor emission spectra). At least two or three experiments were performed for each experimental data, and the error bars are plotted as one standard deviation if there are three data points, or as the offset between each data and the average if there are two data points. The EFRET was calculated by using the (ratio)A method of normalization for the acceptor fluorescence.[31] The EFRET is inversely proportional to the sixth power of the distance between the fluorophores:
in which R is the distance between the donor and acceptor and R0 is the Förster length (the distance at which the energy transfer efficiency is 50%) for the two fluorophores used.
Supplementary Material
Acknowledgements
We thank Dr. Nesha May Andoy and Dr. Peng Chen from Cornell University for helpful discussions about FRET data fitting, Dr. Nandini Nagraj for helpful discussions about data interpretation, and Hannah E. Ihms for carefully proofreading of the manuscripts. This work has been supported by the Office of Science (BER), the U.S. Department of Energy (DE-FG02-08ER64568), the U.S. National Institutes of Health (ES16865), and the U.S. National Science Foundation (CTS-0120978 and DMI-0328162).
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.201100352.
References
- 1.a) Jamieson ER, Lippard SJ. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]; b) Erkkila KE, Odom DT, Barton JK. Chem. Rev. 1999;99:2777–2796. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]; c) Lu Y. Chem. Eur. J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]; d) DeRose VJ. Curr. Opin. Struct. Biol. 2003;13:317–324. doi: 10.1016/s0959-440x(03)00077-0. [DOI] [PubMed] [Google Scholar]; e) Lippert B. Prog. Inorg. Chem. 2005;54:385–447. [Google Scholar]; f) Jiang Q, Xiao N, Shi PF, Zhu YG, Guo ZJ. Coord. Chem. Rev. 2007;251:1951–1972. [Google Scholar]; g) Sigel RKO, Pyle AM. Chem. Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]; h) Bruijnincx PCA, Sadler PJ. Curr. Opin. Chem. Biol. 2008;12:197–206. doi: 10.1016/j.cbpa.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Reedijk J. Platinum Met. Rev. 2008;52:2–11. [Google Scholar]; j) Sigel RKO, Sigel H. Acc. Chem. Res. 2010;43:974–984. doi: 10.1021/ar900197y. [DOI] [PubMed] [Google Scholar]
- 2.a) Manning GS. Acc. Chem. Res. 1979;12:443–449. [Google Scholar]; b) Pan T, Uhlenbeck OC. Nature. 1992;358:560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]; c) Sosnick TR, Pan T. Curr. Opin. Struct. Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]; d) Woodson SA. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]; e) Schnabl J, Sigel RKO. Curr. Opin. Chem. Biol. 2010;14:269–275. doi: 10.1016/j.cbpa.2009.11.024. [DOI] [PubMed] [Google Scholar]; f) Chen P, Andoy NM, Benitez JJ, Keller AM, Panda D, Gao F. Nat. Prod. Rep. 2010;27:757–767. doi: 10.1039/b906691h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]; b) Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 5.a) Sen D, Geyer CR. Curr. Opin. Chem. Biol. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]; b) Silverman SK. Chem. Commun. 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]; c) Schlosser K, Li Y. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Kuwahara M, Sugimoto N. Molecules. 2010;15:5423–5444. doi: 10.3390/molecules15085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santoro SW, Joyce GF. Proc. Natl. Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Faulhammer D, Famulok M. Angew. Chem. 1996;108:2984–2988. [Google Scholar]; Angew. Chem. Int. Ed. Engl. 1996;35:2837–2841. [Google Scholar]; b) Peracchi A. J. Biol. Chem. 2000;275:11693–11697. doi: 10.1074/jbc.275.16.11693. [DOI] [PubMed] [Google Scholar]
- 9.Carmi N, Balkhi SR, Breaker RR. Proc. Natl. Acad. Sci. USA. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Zheng W, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz RPG, Withers JB, Li YF. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc. Natl. Acad. Sci. USA. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. Angew. Chem. 2008;120:4418–4422. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2008;47:4346–4350. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown AK, Li J, Pavot CMB, Lu Y. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- 16.a) Liu J, Lu Y. J. Am. Chem. Soc. 2002;124:15208–15216. doi: 10.1021/ja027647z. [DOI] [PubMed] [Google Scholar]; b) Kim H-K, Liu J, Li J, Nagraj N, Li M, Pavot CMB, Lu Y. J. Am. Chem. Soc. 2007;129:6896–6902. doi: 10.1021/ja0712625. [DOI] [PubMed] [Google Scholar]; c) Lam JCF, Li Y. ChemBioChem. 2010;11:1710–1719. doi: 10.1002/cbic.201000144. [DOI] [PubMed] [Google Scholar]
- 17.a) Kim H-K, Rasnik I, Liu J, Ha T, Lu Y. Nat. Chem. Biol. 2007;3:763–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee NK, Koh HR, Han KY, Kim SK. J. Am. Chem. Soc. 2007;129:15526–15534. doi: 10.1021/ja0725145. [DOI] [PubMed] [Google Scholar]
- 18.a) Liu Y, Sen D. J. Mol. Biol. 2008;381:845–859. doi: 10.1016/j.jmb.2008.06.036. [DOI] [PubMed] [Google Scholar]; b) Liu Y, Sen D. J. Mol. Biol. 2010;395:234–241. doi: 10.1016/j.jmb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 19.a) Dahm SC, Uhlenbeck OC. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]; b) Bassi GS, Murchie AIH, Walter F, Clegg RM, Lilley DMJ. EMBO J. 1997;16:7481–7489. doi: 10.1093/emboj/16.24.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Proc. Natl. Acad. Sci. USA. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Draper DE. Biophys. J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown AK, Liu J, He Y, Lu Y. ChemBioChem. 2009;10:486–492. doi: 10.1002/cbic.200800632. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Wang Z, Liu J, Lu Y. J. Am. Chem. Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang Y, Wang ZD, Xing H, Wong NY, Lu Y. Anal. Chem. 2010;82:4122–4129. doi: 10.1021/ac100244h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moshe M, Elbaz J, Willner I. Nano Lett. 2009;9:1196–1200. doi: 10.1021/nl803887y. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Lu Y. Angew. Chem. 2007;119:7731–7734. [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:7587–7590. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]
- 25.a) Murray JB, Terwey DP, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott WG. Cell. 1998;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]; b) Martick M, Scott WG. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammann C, Cooper A, Lilley DMJ. Biochemistry. 2001;40:1423–1429. doi: 10.1021/bi002231o. [DOI] [PubMed] [Google Scholar]
- 27.a) Zacharias M, Hagerman PJ. J. Mol. Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]; b) Orr JW, Hagerman PJ, Williamson JR. J. Mol. Biol. 1998;275:453–464. doi: 10.1006/jmbi.1997.1489. [DOI] [PubMed] [Google Scholar]
- 28.a) Walter NG, Harris DA, Pereira MJB, Rueda D. Biopolymers. 2002;61:224–242. doi: 10.1002/bip.10144. [DOI] [PubMed] [Google Scholar]; b) Pan T, Sosnick T. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]; c) Lilley DMJ. Methods Enzymol. 2009;469:159–187. doi: 10.1016/S0076-6879(09)69008-X. [DOI] [PubMed] [Google Scholar]
- 29.a) Vogt M, Lahiri S, Hoogstraten CG, Britt RD, DeRose VJ. J. Am. Chem. Soc. 2006;128:16764–16770. doi: 10.1021/ja057035p. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kisseleva N, Kraut S, Jäschke A, Schiemann O. Hfsp. J. 2007;1:127–136. doi: 10.2976/1.2756332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.a) Sigel H, Griesser R. Chem. Soc. Rev. 2005;34:875–900. doi: 10.1039/b505986k. [DOI] [PubMed] [Google Scholar]; b) Johannsen S, Korth MMT, Schnabl J, Sigel RKO. Chimia. 2009;63:146–152. [Google Scholar]; c) Manoharan V, Fürtig B, Jäschke A, Schwalbe H. J. Am. Chem. Soc. 2009;131:6261–6270. doi: 10.1021/ja900244x. [DOI] [PubMed] [Google Scholar]
- 31.Clegg RM. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 32.a) Tuschl T, Gohlke C, Jovin TM, Westhof E, Eckstein F. Science. 1994;266:785–789. doi: 10.1126/science.7973630. [DOI] [PubMed] [Google Scholar]; b) Bassi GS, Murchie AIH, Lilley DMJ. RNA. 1996;2:756–768. [PMC free article] [PubMed] [Google Scholar]; c) Bassi GS, Mollegaard NE, Murchie AIH, Lilley DMJ. Biochemistry. 1999;38:3345–3354. doi: 10.1021/bi982985r. [DOI] [PubMed] [Google Scholar]; d) Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DMJ. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Boots JL, Canny MD, Azimi E, Pardi A. RNA. 2008;14:2212–2222. doi: 10.1261/rna.1010808. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) De Silva C, Walter NG. RNA. 2008;15:76–84. doi: 10.1261/rna.1346609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Zhao ZY, Wilson TJ, Maxwell K, Lilley DMJ. RNA. 2000;6:1833–1846. doi: 10.1017/s1355838200001230. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wilson TJ, Lilley DMJ. RNA. 2002;8:587–600. doi: 10.1017/s1355838202020514. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wilson TJ, Nahas M, Araki L, Harusawa S, Ha T, Lilley DMJ. Blood Cells Mol. Dis. 2007;38:8–14. doi: 10.1016/j.bcmd.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 34.a) Pereira MJB, Harris DA, Rueda D, Walter NG. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]; b) Harris DA, Rueda D, Walter NG. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]; c) Jeong S, Sefcikova J, Tinsley RA, Rueda D, Walter NG. Biochemistry. 2003;42:7727–7740. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]; d) Tinsley RA, Harris DA, Walter NG. Biochemistry. 2004;43:8935–8945. doi: 10.1021/bi049471e. [DOI] [PubMed] [Google Scholar]; e) Gondert ME, Tinsley RA, Rueda D, Walter NG. Biochemistry. 2006;45:7563–7573. doi: 10.1021/bi052116j. [DOI] [PubMed] [Google Scholar]
- 35.a) Lafontaine DA, Wilson TJ, Norman DG, Lilley DMJ. J. Mol. Biol. 2001;312:663–674. doi: 10.1006/jmbi.2001.4996. [DOI] [PubMed] [Google Scholar]; b) Lafontaine DA, Norman DG, Lilley DMJ. EMBO J. 2001;20:1415–1424. doi: 10.1093/emboj/20.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lafontaine DA, Norman DG, Lilley DMJ. Biochimie. 2002;84:889–896. doi: 10.1016/s0300-9084(02)01406-2. [DOI] [PubMed] [Google Scholar]; d) Lafontaine DA, Norman DG, Lilley DMJ. EMBO J. 2002;21:2461–2471. doi: 10.1093/emboj/21.10.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Pereira MJB, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. J. Mol. Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.a) Steiner M, Karunatilaka KS, Sigel RKO, Rueda D. Proc. Natl. Acad. Sci. USA. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Steiner M, Rueda D, Sigel RKO. Angew. Chem. 2009;121:9920–9924. doi: 10.1002/anie.200903809. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:9739–9742. doi: 10.1002/anie.200903809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobitski AY, Nierth A, Helm M, Jäschke A, Nienhaus GU. Nucleic Acids Res. 2007;35:2047–2059. doi: 10.1093/nar/gkm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.a) Clegg RM, Murchie AIH, Zechel A, Carlberg C, Diekmann S, Lilley DMJ. Biochemistry. 1992;31:4846–4856. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]; b) Clegg RM, Murchie AIH, Lilley DMJ. Braz. J. Med. Biol. Res. 1993;26:405–416. [PubMed] [Google Scholar]; c) Clegg RM, Murchie AIH, Lilley DMJ. Biophys. J. 1994;66:99–109. doi: 10.1016/S0006-3495(94)80765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Liu J, Declais AC, Lilley DMJ. J. Mol. Biol. 2004;343:851–864. doi: 10.1016/j.jmb.2004.08.079. [DOI] [PubMed] [Google Scholar]; e) McKinney SA, Tan E, Wilson TJ, Nahas MK, Declais AC, Clegg RM, Lilley DMJ, Ha T. Biochem. Soc. Trans. 2004;32:41–45. doi: 10.1042/bst0320041. [DOI] [PubMed] [Google Scholar]
- 39.Clegg RM, Murchie AIH, Zechel A, Lilley DMJ. Proc. Natl. Acad. Sci. USA. 1993;90:2994–2998. doi: 10.1073/pnas.90.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gohlke C, Murchie AIH, Lilley DMJ, Clegg RM. Proc. Natl. Acad. Sci. USA. 1994;91:11660–11664. doi: 10.1073/pnas.91.24.11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grossmann TN, Roeglin L, Seitz O. Angew. Chem. 2007;119:5315–5318. doi: 10.1002/anie.200700289. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:5223–5225. doi: 10.1002/anie.200700289. [DOI] [PubMed] [Google Scholar]
- 42.a) Mergny JL, Maurizot JC. ChemBioChem. 2001;2:124–132. doi: 10.1002/1439-7633(20010202)2:2<124::AID-CBIC124>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]; b) Alberti P, Mergny JL. Proc. Natl. Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mergny JL. Biochemistry. 1999;38:1573–1581. doi: 10.1021/bi982208r. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Lu Y. J. Am. Chem. Soc. 2000;122:10466–10467. [Google Scholar]
- 45.a) Norman DG, Grainger RJ, Uhrin D, Lilley DMJ. Biochemistry. 2000;39:6317–6324. doi: 10.1021/bi992944a. [DOI] [PubMed] [Google Scholar]; b) Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DMJ. Proc. Natl. Acad. Sci. USA. 2008;105:11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Iqbal A, Wang L, Thompson KC, Lilley DMJ, Norman DG. Biochemistry. 2008;47:7857–7862. doi: 10.1021/bi800773f. [DOI] [PubMed] [Google Scholar]
- 46.a) Miyoshi D, Sugimoto N. Biochimie. 2008;90:1040–1051. doi: 10.1016/j.biochi.2008.02.009. [DOI] [PubMed] [Google Scholar]; b) Muhuri S, Mimura K, Miyoshi D, Sugimoto N. J. Am. Chem. Soc. 2009;131:9268–9280. doi: 10.1021/ja900744e. [DOI] [PubMed] [Google Scholar]
- 47.Kazakov SA, Hecht SM. Nucleic Acid–Metal Ion Interactions, Encyclopedia of Inorganic. New York: Wiley; 2006. [Google Scholar]
- 48.Fedor MJ. Annu. Rev. Biophys. Bioeng. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 49.Bassi GS, Moellegaard N-E, Murchie AIH, von Kitzing E, Lilley DMJ. Nat. Struct. Biol. 1995;2:45–55. doi: 10.1038/nsb0195-45. [DOI] [PubMed] [Google Scholar]
- 50.Qu X, Smith GJ, Lee KT, Sosnick TR, Pan T, Scherer NF. Proc. Natl. Acad. Sci. USA. 2008;105:6602–6607. doi: 10.1073/pnas.0801436105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.a) Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]; b) O’Rear JL, Wang S, Feig AL, Beigelman L, Uhlenbeck OC, Herschlag D. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Curtis EA, Bartel DP. RNA. 2001;7:546–552. doi: 10.1017/s1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Perrotta AT, Been MD. Biochemistry. 2006;45:11357–11365. doi: 10.1021/bi061215+. [DOI] [PubMed] [Google Scholar]; e) Kisseleva N, Khvorova A, Westhof E, Schiemann O, Wolfson AD. Oligonucleotides. 2008;18:101–110. [Google Scholar]; f) Lee T-S, Giambasu GM, Sosa CP, Martick M, Scott WG, York DM. J. Mol. Biol. 2009;388:195–206. doi: 10.1016/j.jmb.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Lambert D, Leipply D, Shiman R, Draper DE. J. Mol. Biol. 2009;390:791–804. doi: 10.1016/j.jmb.2009.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazumdar D, Nagraj N, Kim H-K, Meng X, Brown AK, Sun Q, Li W, Lu Y. J. Am. Chem. Soc. 2009;131:5506–5515. doi: 10.1021/ja8082939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.a) Cech TR, Herschlag D, Piccirilli JA, Pyle AM. J. Biol. Chem. 1992;267:17479–17482. [PubMed] [Google Scholar]; b) Fedor MJ. Curr. Opin. Struct. Biol. 2002;12:289–295. doi: 10.1016/s0959-440x(02)00324-x. [DOI] [PubMed] [Google Scholar]
- 54.Geyer CR, Sen D. Chem. Biol. 1997;4:579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- 55.Ke A, Ding F, Batchelor JD, Doudna JA. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 56.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang XW. Proc. Natl. Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.