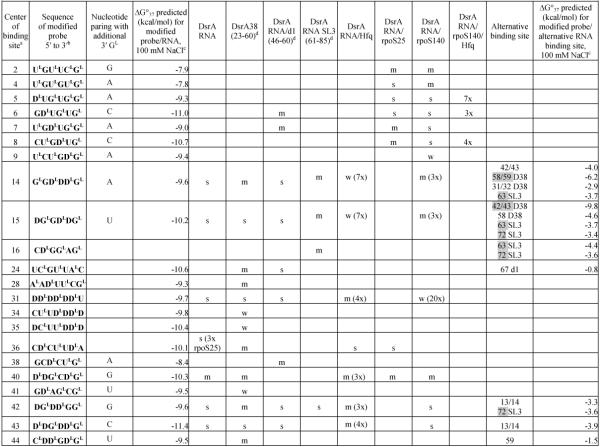
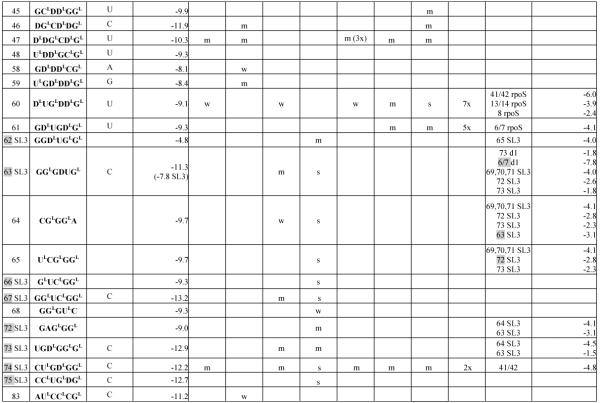
Table 1.
Hybridization results for DsrA RNA and itscomplexes
|
|
Center of binding site is the target RNA nucleotide complementary to the third nucleotide from the 5′-end of the probe.
In sequence of modified probe: LNA nucleotides are marked with superscript L; D represents 2,6-diaminopurineriboside; nucleotides without a superscript are 2′-O-Me-nucleotides.
Calculated according to a published equation (2).
Numbers in brackets show positions in DsrA RNA. Symbols: s - strong binding, m - medium binding, w - weak binding. Numbers with X mean how many times binding signals are diminished: for DsrA/Hfq and DsrA/rpoS140 relative to DsrA alone, for DsrA/rpoS140/Hfq relative to DsrA/rpoS140. The most probably complementary or alternative binding sites are marked grey. If in alternative binding site there are two numbers separated by a slash, it means that probe may bind to an even number of nucleotides.
