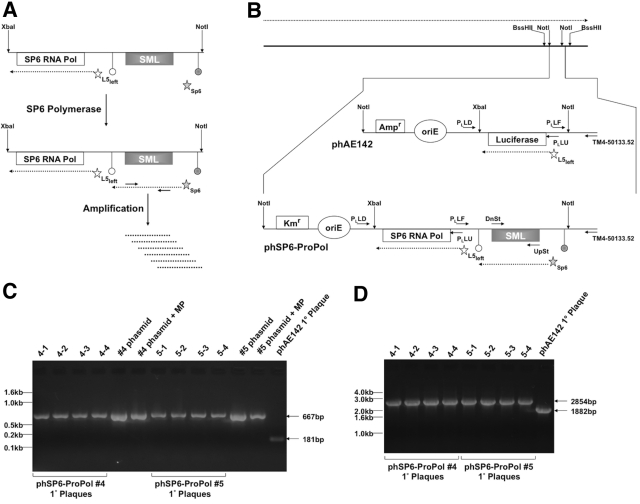
FIG 1 .
Design and characterization of phSP6-ProPol. (A) The SGM was comprised of 2 sections contained within a XbaI-NotI restriction fragment: the SP6 RNA polymerase gene (SP6 RNA Pol) under transcriptional control of the mycobacteriophage L5 Pleft promoter (open star) and the consensus SP6 promoter fused to the SML-encoding sequence (filled star). The SP6-SML section is flanked by 2 transcription terminators: the upstream terminator (filled circle) is E. coli rrnBT2 and precludes basal transcription through the SML-encoding sequence by host RNA polymerase; the downstream terminator (open circle) is the SP6 RNA polymerase terminator from the region downstream of the SP6 phage major capsid subunit described by Dobbins et al. (22). After expression of SP6 RNA polymerase from Pleft, the SML-encoding sequence downstream of the SP6 promoter was transcribed by SP6 RNA polymerase. SP6-dependent transcription of the SML-encoding sequence constituted generation of the SML. SML RNA could then be amplified and detected using primers that bind the SML. (B) The TM4 genome is depicted by the solid black line at the top of the figure. Expression of phage genes occurs on one strand of the genome, and the direction of transcription is indicated by the dashed arrow above the phage genome. Transgenic functions inserted into TM4 are contained on a NotI fragment, which is indicated and expanded. phAE142 encodes an ampicillin resistance cassette (Ampr) and an origin of replication (oriE) for maintenance and selection of the phasmid in E. coli. phAE142 also encodes the luciferase open reading frame fused to Pleft. phSP6-ProPol was derived from phAE142 and replaced the luciferase-encoding XbaI-NotI fragment with the XbaI-NotI SGM. In addition, phSP6-ProPol contained a kanamycin resistance cassette (Kmr) in place of phAE142 Ampr. Pleft transcription occurred on the strand opposite the endogenous phage functions in both phAE142 and phSP6-ProPol. The binding sites and orientation of oligonucleotide primers Ul53-UpSt-113348 and Ul53-DnSt-112112 (UpSt and DnSt, respectively) used for detection of SML generation and those used to characterize transgene structure in phSP6-ProPol are indicated. (C) Phage eluted from primary (1°) plaques originating with transformation of 2 independent phSP6-ProPol-Kan phasmid DNA clones (#4 and #5), as well as phAE142 phasmid DNA, into mc2 4502 were added to a PCR with the primers PLLF and TM4-50133.52. Phasmid DNA with and without the addition of MP buffer was included as controls. PLLF and TM4-50133.52 were predicted to generate a 667-bp product using phSP6-ProPol as a template, compared to a 181-bp product when phAE142 was the substrate. Products were separated on a 2% agarose gel and visualized by ethidium bromide staining. Locations of DNA size markers are indicated. (D) Phage eluted from the 1° plaques in panel C were amplified using primers PLLU and PLLD, which were predicted to mediate amplification of 2,854-bp and 1,882-bp products in phSP6-ProPol and phAE142, respectively. Products were then separated on a 1% agarose gel and visualized by ethidium bromide staining. The locations of DNA size markers are indicated.