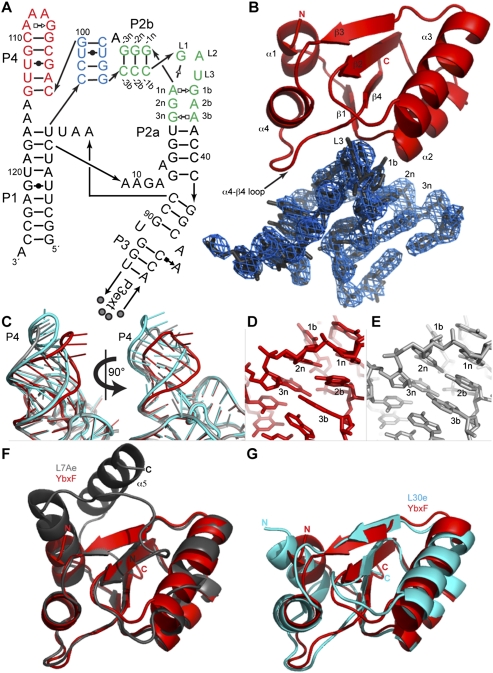
FIGURE 2.
RNA recognition by YbxF. (A) Secondary structure of the Thermoanaerobacter tengcongensis SAM-I riboswitch construct used for cocrystallization. The distal end of P3 was engineered with the P3ext sequence described in Baird and Ferré-D'Amaré (2010). Base pairs observed in the cocrystal structure of the riboswitch bound to YbxF are denoted using Leontis and Westhof (2001) symbols. Arabic numbers indicate the numbering scheme of the riboswitch aptamer domain. K-turn residues are named using the nomenclature of Liu and Lilley (2006). (Green) K-turn; (red) P4 helix; (blue) pseudoknot. (B) Cartoon representation of YbxF bound to the SAM-I riboswitch K-turn. Secondary structure elements of the protein and several residues of the K-turn are named as in Figure 1A and part A, respectively. A portion of the 2.8 Å resolution anneal-omit 2|Fo| − |Fc| electron density map corresponding to the K-turn is shown contoured at 2σ. (C) Superposition of the SAM-I riboswitch core (red, from RNA complexed to YbxF), the protein-free T. tencongensis riboswitch (Montange et al. 2010) (PDB ID 3GX5, gray), and the B. subtilis yitJ riboswitch (Lu et al. 2010) (PDB ID 3NPB, cyan) demonstrates that P4/L4 is mobile in relation to the body of the aptamer domain. (D) Magnified view of the NC helix from the RNA complexed to YbxF reveals no interactions between G at position 2n and A at position 2b. (E) The protein-free SAM-I riboswitch (3GX5) has the same sequence as in D and indicates formation of the G•A sheared pair between 2b–2n residues of the NC helix. (F) Structural alignment of YbxF (red) and L7Ae (gray). (G) Structural alignment of YbxF (red) and L30e (cyan).