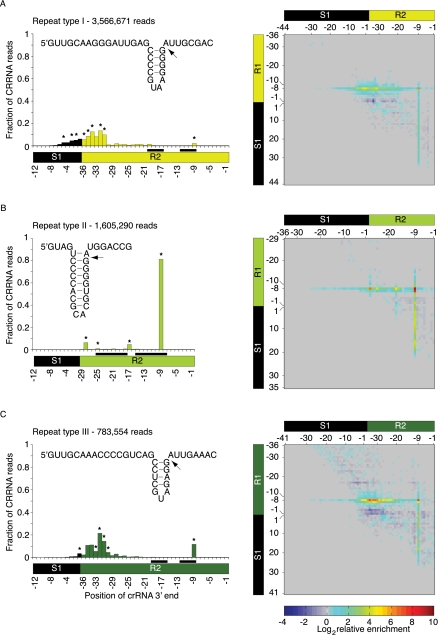
FIGURE 5.
Boundaries of the mature crRNAs can be revealed from the deep-sequencing data. (A–C, left) Average fraction of reads ending at each position of the aligned CRISPR units (for the alignment scheme and the numbering of the aligned positions, see Materials and Methods; Fig. 4A, right). All reads start at the −8 of their respective repeat, annotated as R1. Positions to which a negligible number of read ends map are not displayed. (Inset) Predicted secondary structure models of the three repeat-sequence types. Models were built by identifying the longest and most stable contiguously base-paired region and supporting it by nucleotide covariation that maintains the putative stem (Supplemental Fig. S3). (Arrow) Cleavage site (−8). In repeat-sequence type II, the long stem–loop involves pairing of the cleavage site. Interestingly, this base pair was found to be less stable when complexed with T. Thermophilus HB8 Cse3 (Sashital et al. 2011). Positions predicted to base pair in the secondary structure models (insets) are marked by a pair of black rectangles on the spacer-repeat unit scheme. The number of reads in the analysis is indicated at the top left corner. (A–C, right). Heatmap representation of the average enrichment scores (log2 values) of all the reads starting at the position denoted by the y-axis and ending at the position denoted by the x-axis. Position annotation follows the Figure 4B description except for the spacer start position in the y-axis that is numbered from 1 to N and not from −N to −1. Highly enriched end positions starting at R1–8 (enrichment score is among the top 10 scores of all end positions for each type) are marked in black asterisks in the corresponding left panel graph. For all three repeat-sequence types, the highest average enrichment score was found for reads starting at R1–8 and ending at the R2–9 positions, where the enrichment score of repeat-sequence type II>type III>type I (9.27, 7.36, and 6.48, respectively). High enrichment scores were also observed for fragments starting at −7 positions, predominantly in CRISPRs of repeat-sequence type II fragments starting at R1–7 and ending at R2–9.