FIGURE 5.
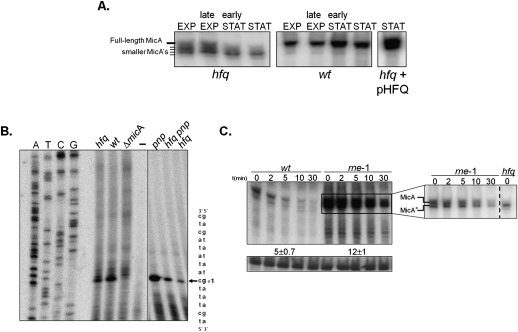
Hfq is required for the expression of the full-length MicA RNA. (A) Steady-state levels of MicA RNA along the growth curve. Culture samples of wild-type or hfq mutant bacteria were collected at exponential (EXP), late exponential, early stationary, and stationary phase (STAT) (corresponding to OD600 values of ∼0.3, ∼1.7, ∼2.5, and ∼5.5 for the wild-type and ∼0.3, ∼0.8, ∼1.6, and ∼2.3 for the hfq mutant, respectively). The growth curves for the wild-type and the hfq mutant strain are given in Supplemental Figure S2. A specific antisense MicA riboprobe was used to detect MicA. Stationary-phase cultures of the hfq mutant transformed with the overexpressing pHFQ plasmid show complementation and do not exhibit the heterogeneous population of MicA's typically found in the hfq single mutant. (B) Determination of the 5′-end of MicA. Total RNA from stationary-phase cells of wild-type, hfq, pnp, and hfq pnp strains was analyzed by primer extension with the [32P]-labeled primer MicA-PE. The same primer extension product (indicated by an arrow) is detected on all strains and absent from the deletion micA strain (ΔmicA) and the negative control reaction (−) done without RNA. Part of the DNA sequence is indicated on the right. The transcription start site of MicA is indicated (+1) and is identical to the site described by Udekwu et al. (2005). The intensity of the primer extension product obtained is higher in the wild-type rather than the hfq mutant, in agreement with the higher amount of MicA detected in the wild-type strain (see Fig. 5A). (C) Northern blot detection of MicA in stationary-phase cultures of Hfq+ cells upon inactivation of RNase E. Cultures of wild-type and an RNase E mutant strain were grown at 30°C until they reached stationary phase, and then shifted to the nonpermissive temperature of 44°C. After 5 min, transcription was blocked with the addition of rifampicin, and samples were withdrawn at times indicated. A specific riboprobe was used to detect MicA RNA. A nonspecific band that cross-hybridized with the antisense MicA probe was used as loading control. The inset corresponds to a shorter exposure of the membrane in which it is visible that both the full-length MicA and the shorter MicA* RNA are detected and stabilized upon inactivation of RNase E in Hfq+ cells. The hfq mutant was used here as a control to clearly identify MicA* RNA.