Abstract
Viruses often induce signaling through the same cellular cascades that are activated by damage to the cellular genome. Signaling triggered by viral proteins or exogenous DNA delivered by viruses can be beneficial or detrimental to viral infection. Viruses have therefore evolved to dissect the cellular DNA damage response pathway during infection, often marking key cellular regulators with ubiquitin to induce their degradation or change their function. Signaling controlled by ubiquitin or ubiquitin-like proteins has recently emerged as key regulator of the cellular DNA damage response. Situated at the interface between DNA damage signaling and the ubiquitin system, viruses can reveal key convergence points in this important cellular pathway. In this review, we examine how viruses harness the diversity of the cellular ubiquitin system to modulate the DNA damage signaling pathway. We discuss the implications of viral infiltration of this pathway for both the transcriptional program of the virus and for the cellular response to DNA damage.
Keywords: ubiquitin, ubiquitin ligase, DNA damage, DNA repair, virus infection
1 Introduction
The DNA damage response (DDR) consists of a coordinated network of proteins that mediate recognition of damaged lesions, activate signal transduction pathways that alter cell cycle progression, and initiate processes that repair the damaged DNA [1,2]. Post-translational modifications, including phosphorylation, acetylation, methylation, SUMOylation and ubiquitination, are utilized to regulate protein function in the damage response [3–5]. These modifications alter protein function, change protein stability, mediate protein-protein interactions, and participate in recruitment of proteins to specific locations during the damage response. In addition to DNA double strand breaks (DSBs) in the cellular genome, viruses also activate the DDR [6,7], and the presence of viral proteins can lead to dramatic changes to the pattern of these post-translational modifications in infected cells. Viruses thus provide powerful tools to understand the inner workings of the cell, to identify key cellular regulators of fundamental cellular pathways, and to reveal mechanistic insights into how these pathways can be manipulated. Post-translational modification of proteins by members of the ubiquitin family is emerging as a key factor in the cellular response to DNA damage and the maintenance of genome stability [8,9]. A diverse and dynamic “ubiquitin landscape” is found at DSBs and is intimately involved in recognition and signaling of the DNA damage [8,10]. In this review we focus specifically on the interplay between viruses and the DNA damage machinery, and we highlight ways in which viruses exploit ubiquitin modifications to manipulate host cell responses to virus infection and alter the ubiquitin landscape.
1.1 Introduction to ubiquitination and the DDR
Modification with ubiquitin and ubiquitin-like proteins is employed as a reversible signal to regulate a vast array of diverse cellular processes. Covalent attachment of a single molecule of the conserved 76 amino acid ubiquitin protein can alter substrate protein activity and localization. Modular ubiquitin-binding domains (UBDs) that bind monoubiquitin or polyubiquitin chains non-covalently can distinguish between different types of ubiquitin modification [11]. Through these interactions, ubiquitin can be utilized as a marker for regulated degradation, and also for non-proteolytic functions such as altered subcellular localization and signaling [12].
Ubiquitination is achieved through a cascade of enzyme activities provided by the E1, E2 and E3 enzymes [13,14]. The first step in ubiquitination is activation by the ubiquitin activating E1 enzyme, which forms a thioester bond with ubiquitin. Activated ubiquitin is then transferred to the ubiquitin-conjugating enzyme E2. The E3 ligase mediates conjugation of ubiquitin to the substrate through an isopeptide bond formed between the C-terminus of ubiquitin and an acceptor amino acid (normally lysine) in the target protein. A single ubiquitin molecule can be conjugated onto a single residue within the substrate (monoubiquitination), or several residues in a target can each be modified with one ubiquitin molecule (multi-ubiquitination). Long polyubiquitin chains can also be formed linked to a lysine in the substrate. The seven lysine residues in ubiquitin can be used for conjugation and the linkage usage has distinct consequences for the substrate protein. Polyubiquitin chains of K48-linked ubiquitin molecules generally targets the substrate for proteasome-mediated degradation. Specialized ubiquitin-binding domains recognize monoubiquitin or polyubiquitin chains with K63 linkages, which signal for non-degradative functions. Ubiquitination is a dynamic and reversible process, and isopeptidases called deubiquitinating enzymes (DUBs) can specifically cleave ubiquitin linkages.
The cellular DDR is a signal transduction cascade regulated by post-translational modifications [3]. In general, the initial steps of the cascade are marked by phosphorylation and acetylation, while the subsequent steps are characterized by ubiquitination and methylation, with ubiquitin chains playing an important role in amplifying the signal. The primary signal kinases activated by damage are the ataxia telangiectasia mutated protein (ATM), the ATM and Rad3-related kinase (ATR) and the catalytic subunit of DNA-dependent kinase (DNA-PKcs). DSBs in genomic DNA are first recognized and bound by the Mre11/Rad50/Nbs1 (MRN) sensor complex or the Ku70/Ku80 proteins, which recruit ATM or DNA-PKcs, respectively. One of the earliest ATM-induced phosphorylation events occurs at S139 on the histone variant H2AX. The phosphorylated H2AX, referred to as γH2AX, acts as a signal for recruitment of downstream effectors. Phosphorylation marks are recognized by phospho-binding modules, such as the forkhead-associated domain (FHA) and the BRCT domain first described in the C-terminus of BRCA1 [15].
Phosphorylation of damage proteins at DSBs facilitates subsequent recruitment of proteins involved in regulatory ubiquitination. Phosphorylated Mdc1, a checkpoint mediator associated with chromatin at damage sites, is bound by the FHA domain in the RING finger protein RNF8 [16–18]. RNF8 recruits the E2 enzyme Ubc13 to mediate ubiquitination of proteins at the damage site, including histones H2A and H2AX. The integral role of ubiquitination in DSB repair is further highlighted by the fact that RNF8-mediated ubiquitination is necessary but not sufficient for the stable accumulation of downstream DNA repair proteins at DSBs. Conjugated ubiquitin accumulates at damage sites and is recognized by a second ligase, RNF168, which contains motifs interacting with ubiquitin (MIUs) that bind ubiquitin modifications at the DSB site [19,20]. Although the mechanistic details are still being elucidated, it appears that RNF168 acts downstream of RNF8 to enhance deposition of ubiquitin signals on histone substrates at damaged DNA and signaling chromatin remodeling events that expose the H4-K20 dimethyl mark to which the mediator 53BP1 binds [21]. The extent to which different ubiquitin linkages are used is not completely understood. Linkage specific antibodies have demonstrated that K63 accumulates at the break [19,20], and it is likely that many substrates and different linkages will ultimately be demonstrated as part of the ubiquitination cascade. DSBs are eventually repaired by homologous recombination (HR) or non-homologous end joining (NHEJ) pathways.
1.2 Viruses and the DDR
It has recently emerged that many viruses induce signaling through the same cellular cascades that are activated by DNA damage to the cellular genome [6,7]. The DDR is triggered as a response to the activities of viral proteins and by the exogenous DNA delivered and replicated by viruses. Although some aspects of the cellular response may be harnessed and exploited by viruses, other parts may need to be subverted to prevent detrimental outcomes for virus infection. Therefore there is a precise dissection of the cellular pathway during infection, and many viruses express proteins that can manipulate the DDR.
There are multiple reasons why the cellular DNA damage response could have detrimental effects on virus infection and would need to be attenuated to ensure efficient virus growth. These include (1) the impact of signaling on protein function and checkpoint activation, (2) the recruitment of repressive factors, and (3) the processing of viral genomic DNA. Since many viruses use the host cell machinery for crucial steps in protein production, such as transcription, splicing, RNA transport and translation, damage signaling that disrupts these cellular processes will have an adverse effect on virus production. Activation of damage signaling also leads to arrest at cell cycle checkpoints which may need to be overcome. Just as accumulation of cellular signaling and repair proteins at DSBs in cellular chromatin leads to silencing of gene expression and processing of DNA ends, recruitment of DDR proteins to viral genomes can impact viral gene expression and DNA replication. Damage recognition leads to histone modifications that signal recruitment of repressive complexes, which suppress gene expression, providing an intrinsic host barrier that must be overcome during the initial steps of infection or reactivation from latent state. Processing of viral genomes by the cellular DNA repair machinery can limit virus replication. DNA resection at the ends of viral genomes can interfere with replication from terminal located viral origins. Processing can also result in ligation or recombination at ends to form concatemers of viral genomes that are too large to be packaged into virus particles. The DNA sequences at the termini of viral genomes must therefore be protected from processing and erosion. To overcome these intrinsic barriers, viruses have evolved multiple strategies to exploit or counter the effects of the cellular DNA damage machinery. Many of these approaches involve hijacking the host ubiquitin-proteasome system to induce targeted degradation of key effectors of the cellular DDR.
1.3 Using ubiquitination to manipulate the DDR
A number of excellent recent reviews have summarized ways in which viruses interact with the ubiquitin-proteasome system (UPS) to blunt immune responses and promote virus replication [22–26]. Since the E3 ligases confer the majority of the substrate specificity to the ubiquitination reaction cascade, it is normally this step that is harnessed by viral proteins to target host factors. There are two main types of E3 ligases: those with a HECT domain (homologous to E6-associated protein C-terminus), and those with RING (really interesting new gene) finger domains. The HECT E3s have direct catalytic activity, whereas the RING E3s act more like scaffolds to bring the E2 and substrates together to facilitate ubiquitination. There are examples of virally-encoded proteins that mediate ubiquitination via HECT domains, such as the E6 protein of human papillomavirus (HPV), which functions with the prototypical E6-associated protein E6-AP [26]. There are also viral ligases that function via RING finger domains, such as the immediate early protein ICP0 of Herpes Simplex Virus type 1 (HSV-1). The RING finger is required for the E3 ligase activity of ICP0 and its substrates can be targeted through direct interactions or via other post-translational modifications (see below). ICP0 uses UbcH5a or UbcH6 as its E2s [27], a selection that appears to be specific as a number of other E2s are unable to substitute. Interestingly, these two E2 proteins were originally identified as facilitating the HPV E6–E6AP-induced degradation of p53.
The Cullin-RING ligases (CRLs) form the largest class of cellular ubiquitin ligases and they consist of multisubunit complexes characterized by an enzymatic core that contains a Cullin family member together with a RING finger protein [28]. Many viruses encode adaptors that assemble with cellular CRLs into ligases that are redirected towards novel substrates [29]. For example, the Adenovirus (Ad) E4orf6/E1b55K proteins assemble into a complex that targets cellular proteins for ubiquitination and degradation [30,31]. The E4orf6 protein recruits the Cullin complex through an interaction with Elongins B and C [32], while the viral E1b55K protein serves as the substrate recognition component [33]. Interestingly, there is heterogeneity in the composition of the E3 ligases assembled by the E1b55K/E4orf6 proteins from different Ad serotypes, with either Cul2 or Cul5 being utilized [34]. In some cases the E4orf6 protein alone can act as a CRL substrate adaptor, binding directly to the substrate and targeting it for degradation in the absence of E1b55K [35].
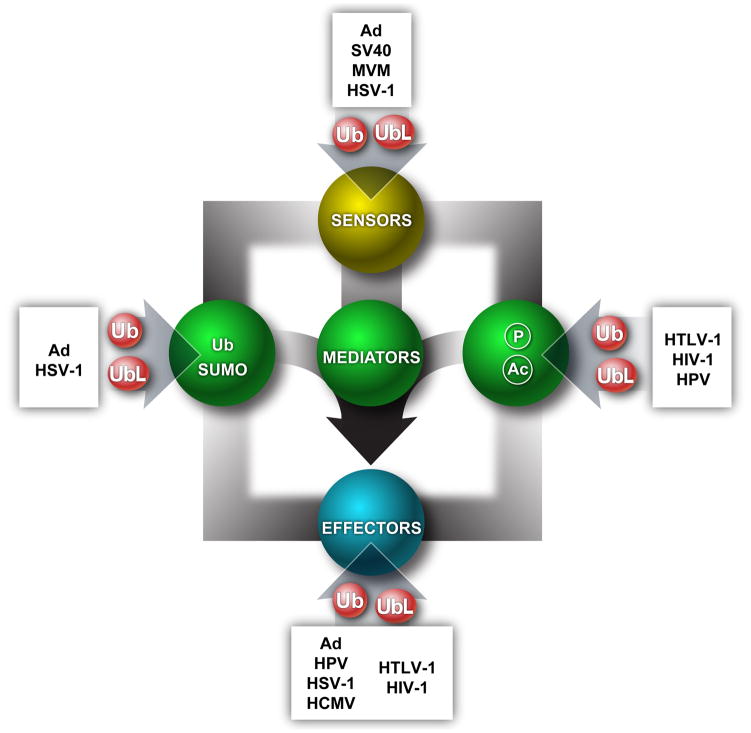
2 Selecting key targets in cellular damage pathways
As intracellular parasites with small genome sizes, viruses have often evolved to target key convergence points in the cellular pathways that they disrupt or exploit. In this way, examining the point at which viruses intervene can identify hubs or reveal important regulators of cellular functions. In the following sections we discuss several of these regulators in the DDR pathway that are targeted by viral ubiquitin ligases (summarized in Figure 1). Although viruses can target common host pathways, they may adopt divergent mechanisms to neutralize the host defenses in order to promote efficient virus growth. Even amongst closely related viruses with a single virus family, different strategies may be employed to target and inactivate DDR pathways and counteract cellular responses. In some cases, in addition to beneficial consequences for the virus, this targeting has consequences for cellular DNA damage recognition and non-degradative ubiquitin signaling.
Figure 1. Viral interfaces with key nodes of the DNA damage signaling pathway.
The cellular response to DNA damage is initiated by proteins that sense the damage and signal to mediator proteins. Mediator proteins reinforce the signaling by a cascade of post-translational modifications including phosporylation, acetylation, ubiquitination, and SUMOylation, and there is extensive crosstalk between these modifications. Together, signaling from these DNA damage response proteins coordinate cell cycle arrest and DNA repair. All of these nodes are targeted by viruses, underscoring the importance of gaining control of this pathway. Studying where viruses interface with the DNA damage response will likely reveal new nodes or regulators of this important cellular pathway.
Abbreviations: Herpes Simplex Virus type 1 (HSV-1), Human T-cell lymphotropic virus (HTLV-1), Simian Virus 40 (SV40), Human Immunodeficiency Virus 1 (HIV-1), Adenovirus (AAV), Minute Virus Mice (MVM), Human Papillomavirus (HPV), Human Cytomegalovirus (HCMV), Hepatitis C Virus (HCV)
2.1 The MRN complex
The MRN complex is composed of proteins known as Mre11, Rad50, and Nbs1. The MRN complex has numerous enzymatic functions and plays a central role in the cellular DDR [36,37]. It has been demonstrated to serve as a sensor of DSBs and is involved in recruitment and activation of ATM at damage sites. The MRN proteins bind to DNA and the complex can bridge DNA ends to mediate repair of two molecules. The complex is also involved in pathway choice for DNA repair processes. Homology-directed repair requires resection at DSBs, and this is initiated by the MRN complex, in combination with other cellular nucleases [38]. MRN is also involved in the resection-dependent alternative NHEJ pathway that is characterized by large deletions and the use of short microhomologies.
The first example of the MRN complex being a target for viral manipulation came from our observation that levels of all three proteins are reduced during infection with Ad5 [39]. We demonstrated that the E1b55K/E4orf6 complex was necessary and sufficient, and that degradation was due to proteasome-dependent turnover [39,40]. The E1b55K protein binds to the MRN complex, and mutants can separate degradation of MRN from targeting of other substrates [40–42]. Degradation is reduced by a trans-dominant negative Cul5 or by shRNA to Cul5, suggesting that the E3 ligase recruited contains this cullin [34,43]. Ubiquitination has not actually been demonstrated on any of the MRN components and it is difficult to determine which complex member is the direct target. Since the complex is unstable in the absence of any single protein, removing any one member may be sufficient to inactivate its functions. Infections in mutant cell lines suggest that Mre11 may be the degradation substrate for the E1b55K/E4orf6 complex of Ad5 [41]. Although most of the Ad serotypes can degrade MRN, this may not be a conserved target of E1b55K/E4orf6 across all the family members [34,44].
We have proposed that Ad degrades the MRN complex in order to prevent its detrimental effects on processing of the viral genome. MRN restricts replication of the viral DNA genome [45–47] and is required for joining viral genomes into concatemers [39]. Since multiple functions are attributed to the MRN complex, its depletion through viral-targeted ubiquitination has a number of consequences for functioning of the host cell DNA damage response. We have shown that depletion of MRN by viral proteins reduces the ability to activate ATM and ATR signaling pathways, and blunts checkpoint activation [40,48]. However, not all signaling events are blocked during virus infection, and there are some Ad serotypes that generate both ATM and ATR dependent phosphorylation signaling [44]. The importance of Ad gaining control of DNA damage signaling is underscored by the fact that redundant or alternative viral strategies also exist, some of which also employ the cellular UPS. For example the E4orf6 protein of the Ad12 serotype recruits a Cul2/Rbx1/elonginC complex to promote proteasomal degradation of TopBP1, which results in inhibition of ATR activation [35].
The central role of MRN in sensing and responding to DNA damage or aberrant DNA structures is highlighted by the number of viruses that target the cellular complex [7]. In addition to Ad, the levels of the MRN complex are also reduced at late times of infection for other viruses. In the case of infection with Simian Virus Type 40 (SV40), the viral T-antigen (LT) recruits a Cul7-containing ubiquitin ligase to facilitate the degradation of the Mre11 complex [49], while Minute Virus of Mouse (MVM) and HSV-1 initially recruit MRN proteins into viral replication compartments but later induce Mre11 degradation [50,51].
2.2 The ligases RNF8 and RNF168
RNF8 is a RING finger ubiquitin ligase that also contains a phospho-binding FHA domain. This combination of motifs allows RNF8 to serve as a molecular adaptor between phosphorylation- and ubiquitination-dependent DNA damage signaling and identifies RNF8 as a central nexus in the DNA damage signaling pathway [16–18,52,53]. The HSV-1 immediate early protein ICP0 degrades RNF8, preventing recruitment of downstream DNA repair proteins to incoming viral genomes or sites of cellular DNA damage [54,55]. RNF8 accumulates at damaged DNA via a phospho-dependent interaction between its FHA domain and ATM-phosphorylated TQXF motifs on the upstream mediator protein, Mdc1. ICP0 has evolved to mimic this interaction by targeting FHA domain of RNF8 via a similar phospho-dependent interaction (M. Chaurushiya et al, unpublished). In the absence of ICP0, the catalytic activity of RNF8 combined with the E2 conjugating enzyme Ubc13 results in the ubiquitination of histone substrates such as H2A and H2AX and these act as a platform for the accumulation of downstream signaling and repair proteins such as RNF168, BRCA1 and 53BP1 [17–20,53]. In the presence of ICP0, RNF8 is degraded and the levels of ubiquitinated H2A and H2AX are reduced, preventing the stable accumulation of downstream repair factors [54]. Interestingly, ICP0 also targets RNF168 for ubiquitination and degradation, underscoring the importance to the virus of gaining control of the pathway at this point [54]. The fact that ICP0 degrades both RNF8 and RNF168 raises the possibility that there may be separate RNF8- and RNF168-dependent functions. For example, ICP0-mediated degradation of RNF168 could guard against amplification of a mono-ubiquitination signal established by an RNF8-independent mechanism. Interestingly, recent studies on transcriptional silencing in the DDR (see below) reveal that transcription in the presence of a DSB requires the simultaneous depletion of both RNF8 and RNF168, suggesting that the ligases are not redundant in this pathway, and hinting at a possible reason ICP0 may need to degrade both proteins.
2.3 Targets that affect chromatin and impact viral gene expression
Upon entering a cell, the primary objective of a virus is to set up a favorable cellular environment for replication, and this is requires efficient transcription from the viral genome. Gaining control of the cellular transcription machinery and defusing any cellular silencing mechanisms is therefore key to establishing a successful infection. It is well recognized that transcription is often regulated by chromatin modifications, and there is now evidence that chromatin changes in response to DNA damage have implications for transcription. Since chromatinization plays an important role in regulating gene expression from viral genomes [56], viral interactions with the DDR may contribute to hijacking or avoidance of cellular transcriptional programs.
The Tip60 tumor suppressor is a histone acetyltransferase involved in transcriptional regulation, checkpoint activation, and apoptosis [57]. It is recruited to DSBs [58] and is required for the mobilization of γH2AX immediately after break induction. Tip60 acetylates H2AX on K5, which promotes ubiquitination events on H2AX, facilitating release of γH2AX from chromatin and signaling termination of the DDR [59,60]. Tip60 forms a stable complex with ATM and acetylates it in response to damage, facilitating ATM activation by auto-phosphorylation [61]. Several viruses have been shown to interact directly with Tip60 [62,63]. Most recently, Tip60 has been found to bind to HPV promoters where it acetylates histone H4, recruiting the cellular protein Brd4 to repress HPV E6 expression [64]. The HPV E6 protein responds by targeting Tip60 for proteasome-mediated degradation, resulting in the de-repression of viral promoters [64].
As described above, the ligases RNF8 and RNF168 are chromatin-modifying enzymes that are involved in the DDR. The contributions of RNF8, RNF168 and uH2A to transcriptional repression at DSBs were recently analyzed using a fluorescent transcriptional reporter system [65]. In this system, DSBs generated in the proximity of a reporter cassette using a targeted nuclease resulted in transcriptional repression that was dependent on ATM, RNF8 and RNF168 and facilitated by ubiquitinated H2A. Interestingly, the authors of this paper speculated that the DSB-induced transcriptional silencing pathway may have evolved to defend the genome against the uncontrolled transcription of foreign genomes resembling DSBs, such as viruses [66]. Other links between RNF8 and transcription come from studies with RNF8 null mice, where male infertility results from loss of chromatin ubiquitination and defects in global nucleosome removal [67]. This defect in nucleosome removal was found to be attributable to a loss of H4K16 acetylation, a histone modification with known roles in transcriptional activation and the maintenance of euchromatin. These findings highlight the potential consequences of RNF8 loss on crosstalk between transcriptionally important histone modifications.
Recent findings have strengthened the links between DNA damage, H2A ubiquitination, and gene silencing, with implications for their manipulation by viruses. Although there is a clear contribution of RNF8 and RNF168 to H2A ubiquitination, the deposition of uH2A as a repressive histone mark is more canonically catalyzed by the polycomb group complex (PRC1) proteins Bmi1 and Ring1b/RNF2/RING2. Recent work has identified Bmi1 as a DNA repair protein, and demonstrated that Bmi1 acts as an adaptor for targeting RING2, the catalytic subunit of the heterodimer, to the sites of damage [68,69]. The PRC1 complex is required for post-damage mono-ubiquitination of H2A, and contributes to homology-mediated repair of DNA breaks. While one of these studies reported that recruitment of Bmi1 to sites of DNA damage was dependent on RNF8 [68], the other suggested that Bmi1 and RNF8 acted in parallel pathways to control H2A ubiquitination [69]. These observations highlight a potentially important parallel between viral control of the processes of damage responses and transcription. Since mono-ubiquitinated H2A is linked to gene silencing and is enriched in silenced chromatin, its manipulation by HSV-1 ICP0 via degradation of RNF8 and RNF168 may represent an example of a virus interfacing with DDR proteins in order to control transcription. In animal models, ICP0 is required for efficient exit from latency [70,71] and in tissue culture systems, mutant viruses unable to express ICP0 can be transcriptionally silenced and persist for extended periods of time [72,73]. These quiescent viral genomes are not permanently inactivated, since they can be reactivated by superinfection with wild-type HSV or by providing ICP0 in trans [72–74]. This suggests that ICP0 controls a reversible silencing mechanism, and it is possible that degradation of RNF8 and RNF168, together with the potential resultant loss of Bmi1 recruitment and uH2A, provides a functional link from cellular mechanisms of transcriptional silencing to ICP0 roles in reactivation. Relevant to this discussion is the recent observation that the PRC1 complex binds to the HSV-1 genome during latency, when ICP0 is not expressed [75].
2.4 The NHEJ machinery to prevent genome processing
Viruses with linear genomes need somehow to prevent joining together of viral genomes. Although the Ad genome is predominantly in a linear monomeric form during infection with wild-type virus, in cells infected with viruses mutated in the early region E4 the viral DNA is joined into end-to-end concatemers [76]. Genomes can be linked to each other in any orientation [76] and analysis of joints reveals heterogeneous terminal deletions with no evidence for the use of microhomology [77]. Concatemer formation requires proteins involved in NHEJ and is not observed in cells with mutant DNA-PKcs and DNA ligase IV [39,78]. The MRN complex is also involved, possibly through processing of the viral ends and removal of a covalently bound terminal protein. During wild-type Ad infection the genome remains linear and therefore the virus expresses proteins that can block the NHEJ-mediated concatemers. In addition to targeting the MRN complex (discussed above), Ad serotype 5 employs a number of other strategies to limit NHEJ. The E1b55K/E4orf6 proteins redirect the cellular Cul5 to assemble an E3 ligase complex to mediate degradation of DNA ligase IV [79]. Prevention of NHEJ activity may be a general requirement across the Ad virus family. Recent studies comparing degradation substrates for the E1b55K/E4orf6 complex from representative serotypes suggest that DNA ligase IV is a conserved target of this E3 ligase [34,44]. This appears sufficient to prevent joining of viral genomes into concatemers by an NHEJ mechanism [44]. In the case of Ad5, it has been shown that Cul5 is involved and the E1b55K protein binds DNA ligase IV directly [79] with a requirement for the BRCT domain (T. Gilson & G. Ketner, personal communication). This highlights an interesting parallel with the ICP0-RNF8 interaction, in which ICP0 binds RNF8 directly via the FHA domain on the substrate. These two examples demonstrate ways that viruses have evolved to mimic post-translational modifications on cellular binding partners of key DDR proteins in order to infiltrate this important cellular pathway.
The activity of the E1b55K/E4orf6 viral complex has a generalized effect on NHEJ. In addition to blocking end-to-end joining of viral genomes, the E1b55K/E4orf6 complex also inhibits NHEJ in other non-viral substrates such as DSBs generated by ionizing radiation, and RAG1/2 generated substrates in a V(D)J plasmid recombination assay [78]. DNA-PKcs and DNA ligase IV have also been proposed to play roles in circularization or processing of HSV-1 genomes. While DNA ligase IV/XRCC4 dependent processing has been proposed to benefit viral replication [80], DNA-PKcs inhibits viral replication and is targeted for degradation by ICP0 [81]. The significance of ICP0-mediated targeting of DNA-PKcs is unclear, although expression of ICP0 inhibited the formation of circular genomes at early times post-infection, raising the possibility that DNA-PKcs may normally serve to circularize incoming viral genomes [82].
2.5 Tumor suppressors p53 and Rb
The classic examples of viruses overtaking key cellular pathways by targeting crucial nodes come from Rb and p53, which together form the crux of G1/S regulation. The Rb family of proteins represses transcription of genes required for entry into S phase by binding and inhibiting the E2F family of transcription factors, while p53 serves as a break on G1/S transition in response to a variety of stress signals, including DNA damage and viral infection. Thus it is imperative for viruses to be able to modulate both Rb and p53 to promote transition to S phase and prevent apoptosis of the infected cell. Canonical studies have described the ability of viruses to inactivate and/or degrade Rb and p53 through protein-protein interactions, and more recent studies have shown that viruses can target E2F and p53-responsive promoters directly to modulate transcriptional events facilitating entry into and through S-phase.
Both Ad-E1A and SV40-LT form stable complexes with Rb, inhibiting its ability to bind and inactivate E2F transcription factors (reviewed in [83]). The E7 protein of HPV and NS5A of hepatitis C virus (HCV) can also degrade Rb through the UPS [84,85], although they each use different methods to ubiquitinate Rb. HPV-E7 assembles a Cul2-based ubiquitin ligase complex [86], while HCV-NS5A recruits the E6AP ubiquitin ligase to target Rb for degradation [87].
These viruses often encode distinct proteins to target p53, or downstream p53 transcriptional targets, to prevent G1/S arrest and promote cell transformation. Degradation of p53 via the UPS is employed by Ad-E1b55K/E4orf6 and HPV-E6, the mechanisms of which have been extensively reviewed [25,88]. More recently it has been shown that viral proteins can also directly target chromatin to modulate transcriptional programs contributing to transformation. Ad-E4orf3 induces heterochromatin formation characterized by H3K9 methylation on p53 target promoters, which prevents p53 binding [89]. Thus even in the presence of stabilized p53, its target genes remain silenced. Viruses can also modulate SUMO pathways to affect the SUMOylation status and downstream functions of p53 (see below).
2.6 The APC and cell cycle
The anaphase-promoting complex/cyclosome (APC) is a multi-subunit E3 ubiquitin ligase complex that acts as a central coordinator of cell cycle, mediating the timely degradation of cyclins and other cell cycle regulators to promote transitions through the different phases [90]. Substrate specificity of the APC is regulated by Cdc20 and Cdh1, which associate with the APC at different times to facilitate transition from G2 and early mitotic events (Cdc20) through anaphase and late mitotic events and back into G1 (Cdh1). For some viruses, it is imperative to maintain the cell in G1/S in order to ensure availability of the cellular DNA replicative machinery. For other viruses, nuclear breakdown due to extended G2/M delay can facilitate nuclear entry of pre-integration complexes for integration into the host genome, or exit of newly synthesized genomes into the cytoplasm for packaging. Therefore the APC/C complex is an attractive target for viruses attempting to modulate different stages of the cell cycle, and different viruses target distinct subunits [91]. The Ad E4orf4 protein recruits an inactivating phosphatase, PP2A, to the complex [92]. The APC has been reported to be deregulated by several mechanisms during human cytomegalovirus (HCMV) infection, including virally-induced phosphorylation of Cdh1 [93], and more recently by degradation of the APC subunits APC4 and APC5 [94]. The APC is also targeted by Tax protein of Human T-cell Lymphotropic Virus (HTLV), the Vpr protein from Human Immunodeficiency Virus (HIV), and the E6/E7 proteins of HPV [91]. The Hepatitis B virus X protein (HBX) dysregulates the mitotic checkpoint by binding BubR1, thereby disrupting the association between BubR1 and the APC activator, Cdc20 [95]. Perhaps most interestingly, a poxvirus protein (PACR, poxvirus APC/C regulator) has recently been shown to mimic the role of the APC11 subunit that promotes its E3 ubiquitin ligase activity [96]. While both APC11 and PACR have similar RING-H2 domains, a specific sequence variation in the PACR RING-H2 domain renders it unable to promote ubiquitin conjugation. PACR can bind to the normal APC11 binding partner APC2, integrating itself into the APC and impairing APC ubiquitin ligase activity. PACR integration into the APC thus leads to accumulation of normal APC substrates and deregulated cell cycle that promotes viral growth. In addition to cell cycle control, APC and Cdh1 also regulate the DNA damage checkpoint response and DNA repair through degradation of a growing list of substrates, linking their activity to maintenance of genomic integrity [97]. Inappropriate regulation of the APC by viral proteins could therefore contribute to transformation and tumorigenesis.
2.7 The DDB1/Cul4 ligase
Cul4 is unique among Cullins in that it is able to form a complex with DDB1, a protein that was identified by its ability to recognize UV-induced DNA damage lesions, and which shares no sequence homology with any known Cullin adaptor [98]. DDB1 has been implicated in regulation of cell proliferation, DNA repair, and genomic integrity through targeted ubiquitination of key cellular regulators. The HIV and simian immunodeficiency virus (SIV) accessory protein Vpr specifically associates with DDB1/Cul4 through an adaptor protein VprBP/DCAF1 (reviewed in Ref [99]). The interaction of Vpr with DDB1/Cul4 is required for a number of the Vpr functions, including G2 cell cycle arrest and activation of the DDR [100,101]. Although some degradation targets have been identified, it is unclear exactly how Vpr exploits DDB1/Cul4 to gain control of the cell cycle and promote viral infection. However, it appears that gaining control of the DDR pathway at the level of the DDB1/Cul4 complex is important for several viruses. Murine gamma herpesvirus 68 (MHV68) latency-associated protein M2 protein inhibits DNA damage-induced apoptosis by interacting with both the DDB1 complex and ATM [102]. Other viral proteins such as paramyxovirus SV5, HBX and woodchuck Hepatitis B virus X protein (WHX) all hijack the DDB1/Cul4 E3 ligase complex, to facilitate replication and promote degradation of proteins that respond to interferon signaling and mediate antiviral responses. Strikingly, structural analysis revealed that these diverse viral proteins bind to DDB1 similarly, possibly by mimicking the cellular DCAF adaptors [103]. These data are an example of how studying conserved viral strategies can pinpoint key cellular regulators. In this case, a structural element that is important for the assembly of both cellular and virally hijacked DDB1/Cul4 E3 complexes was identified [103].
3. Regulating the effect of viral and cellular ubiquitin ligases
The E3 ubiquitin ligases can themselves be modulated by post-translational modifications. There are many examples of cellular ligases that are regulated by phosphorylation, interacting proteins, or conjugation by other ubiquitin-like proteins such as NEDD8. These modifications can promote or prevent intermolecular and intramolecular interactions, or can produce conformation changes that regulate ubiquitination of substrates. E3 ligases can also be regulated at the proteolytic level by degradation, in negative feedback loops induced by self-catalyzed ubiquitination or through the activity of another ligase [104]. Ubiquitin modification on ligases and substrates can also be reversed by the deubiquitinating enzymes or DUBs.
3.1 Manipulating via deubiquitination
Several DUBs have already been implicated in regulating the DDR [8]. Examples include BRCC36 that acts preferentially on K63 linkages and is recruited to DSBs as part of the RAP80 complex [105]. Other DUBs can also reverse chromatin ubiquitination at DSBs, affecting formation of damaged-induced uH2A foci, possibly by counteracting targets of the damage ligases RNF8 and RNF168 [20]. The USP16 enzyme also negatively regulates uH2A, with implications for repression of transcription at DSBs [65]. Another DUB, OTUB1, directly binds and inhibits the E2 Ubc13 by a mechanism independent of its catalytic isopeptidase activity [106]. These examples demonstrate the versatility of DUBs in controlling ubiquitination at DSBs. Viruses could alter both the E3 ligases and their respective DUBs, and thus disrupt the dynamics of ubiquitination at damage sites.
Ligases that undergo auto-ubiquitination can be stabilized through removal of ubiquitin by DUBs. The RING-containing E3 ligase ICP0 from HSV-1 binds to the isopeptidase USP7, and this protects ICP0 from auto-ubiquitination in vitro [107]. There is a reciprocal relationship between these two proteins: their interaction greatly increases the stability of ICP0, while ICP0 also targets USP7 for ubiquitination and proteasome-dependent degradation [108]. The Epstein Barr Virus (EBV) protein EBNA1 also interacts with USP7 [109], suggesting that this isopeptidase may be commonly usurped by viruses. Similar situations may exist for related viral E3 ligases and the balance between the two will have functional consequences for virally-mediated control of ubiquitination in the DDR. For example, USP7 is known to deubiquitinate several emerging and established players in DDR responses and cell cycle regulation including H2B, the H2A ubiquitin ligase RING2, p53, Mdm2 and claspin. Regulation of USP7 by viruses may enable fine-tuning of ubiquitin signals on targeted cellular proteins and may indirectly affect damage responses. For example a recent paper reports that the DNA methyltransferase DNMT1, a co-repressor that is recruited to DSBs where it stimulates DNA methylation, is degraded through acetylation and ubiquitination, which is counteracted by the DUB activity of USP7 [110]. This suggests that viral interactions with USP7 could indirectly have an effect on methylation-dependent silencing mediated by DNMT1. Viruses could also indirectly impact the steady-state levels of ubiquitination on substrates through upregulation of cellular DUBs.
Several viruses encode their own DUBs or are able to increase the activity of cellular DUBs [23]. Viral proteins possessing deubiquitinating activity appear to be particularly common in herpesviruses, perhaps in part due to their larger genome size [111]. In fact, all known herpesviruses sequenced to date contain homologs of the first identified herpesviral DUB, HSV-1 UL36 [112], and in many cases these DUBs have been shown to be required for optimal viral infection. UL36 resides in the tegument layer located between the nucleocapsid and the envelope [113], and is carried into the infected cell, suggesting that candidate substrates may include intrinsic anti-viral defense proteins encountered by the virus at the earliest stages of infection. In the case of herpesviruses, these intrinsic anti-viral defense proteins include ND10 components and certain DNA repair proteins [55,114]. A tegument-delivered viral counter that did not need to be expressed de novo would appear ideally placed to act at this level.
3.2 Manipulating via SUMOylation
Crosstalk between post-translational modifications such as phosphorylation, ubiquitination, and more recently, SUMOylation, has been shown to be important for the successful propagation of a DNA damage response, primarily in mediating protein-protein interactions [3,115]. As described above, viruses have evolved mechanisms to take advantage of both phosphorylation and ubiquitination pathways to modulate the cellular DDR. SUMOylation has been implicated in multiple DNA repair pathways [115], and its manipulation by viral proteins may alter DDR outcomes during infection. SUMO proteins and their ligases accumulate at DSBs in mammalian cells [116], where they promote repair and may increase the activity of ubiquitin ligases [117]. Several viral regulatory proteins and transcriptional transactivators have been shown to be SUMOylated, including the IE1 and IE2 of HCMV [118,119], the BZLF1 and Rta proteins of EBV [120,121], and the E1b55K protein of Ad [122]. While many of these proteins are known to modulate DNA damage responses and/or cell cycle progression, the contributions of SUMOylation to these functions is not as well understood.
Recent descriptions of the first virally-encoded SUMO E3 ligases, both of which can target p53, highlight the importance of infiltrating the SUMO pathway during infection. SUMOylation of transcription factors has been shown both to activate and repress their activities, and examples of both exist in the case of viral SUMOylation of p53. The b-ZIP protein from Kaposi’s Sarcoma Herpes Virus (KSHV) is a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific [123]. KHSV b-ZIP can directly SUMOylate p53 and Rb as well as itself, and this function is critical for its ability to activate the transcriptional capabilities of p53 and induce cell cycle arrest [123]. The Ad E1B55K protein is also a SUMO1-specific E3 ligase that SUMOylates p53 in vitro, and promotes p53 SUMOylation when expressed in cells [124]. This SUMOylation appears to cause p53 to be retained at PML bodies, inhibiting its ability to localize to promoters and activate transcription.
Thus it appears that many viral regulatory proteins, which have previously been described to modulate the DDR, are also connected to the SUMO pathway. It has recently been found that certain proteins known to interface with the DDR pathway can identify targets by recognizing SUMO modifications on potential substrates [125]. One cellular example is RNF4, a SUMO-dependent ubiquitin E3-ligase implicated in maintenance of genomic stability [126,127]. It has recently been found that ICP0, the ubiquitin ligase from HSV-1, encodes SIMs and can target SUMOylated substrates for ubiquitination and degradation (C. Boutell and R. Everett, personal communication). As we have discussed, ICP0 interacts with the DDR pathway at multiple levels, including targeting of DNA-PKcs, RNF8 and RNF168 for degradation. It will be interesting to determine whether the SUMO-targeted ubiquitin ligase activity of ICP0 also enables it to interact with SUMO-modified DDR proteins. We expect future work to reveal that SUMO-based protein-protein interaction mechanisms will be integral for viruses to target the DDR, as well as other SUMO-regulated signaling pathways.
4. Conclusion
Phosphorylation and ubiquitination are reversible post-translational modifications that play important signaling functions in the cellular DDR and are both manipulated by viruses to aid infection. Given the important roles recently revealed for ubiquitin and SUMO in DNA damage and repair pathways, it is likely that many more examples of their modification will come from watching how viruses inactivate the intrinsic antiviral function of the DDR. Most of the examples we have discussed here come from viral exploitation of ubiquitin to target cellular proteins for degradation. It is likely that there will also be many examples of viruses harnessing non-degradative ubiquitin signaling pathways, but so far very few of these have been uncovered.
In addition to repair of DSBs, there are additional wings of the DNA damage network that also involve ubiquitin modifications. These include the Fanconi anemia pathway that is used to repair interstrand crosslinks during replication, and DNA damage tolerance to bypass replicative lesions [9]. Although we have focused on manipulation of ubiquitination by viruses in the DSB sensing and repair pathways, it is likely that links also exist with these other repair systems. For example, it has already been demonstrated that the Fanconi anemia pathway, which is associated with replication stress and results in monoubiquitinated FancD2 retention in chromatin, is activated during Ad infection [128] and by expression of viral oncogenes such as the LT from SV40 and the E7 protein from HPV [129,130]. The replication processivity factor PCNA also gets monubiquitinated after DNA damage and leads to the recruitment of damage-tolerant DNA polymerases required for translesion synthesis. Since PCNA is involved in replication of some viral genomes and is often recruited to viral replication centers, it will be interesting to see whether its post-translational modifications are affected during virus infections.
As with the role of ubiquitination in the DDR in general, questions remain about viral modulation of the DNA damage associated ubiquitination, such as how it is regulated, what the relevant substrates are, and the importance of different ubiquitin chain linkages. The comparison across the Ad serotypes demonstrates that closely related viruses within a single family can adopt different strategies to inactivate DNA damage responses during infection. It will be informative to characterize the full composition of the E3 ligases assembled across a family of viruses, and to determine how this defines the substrates that are targeted. It will also be important to study how modifications render cellular proteins susceptible or resistant to viral-induced ubiquitination.
The function of post-translational modifications is often mediated by specific recognition of the modification by downstream proteins that harbor modular effector domains. Covalent modification by ubiquitin and related proteins changes the surface properties of a protein and thus provides novel interaction sites. Ubiquitin binding domains act as receptors that recognize the ubiquitin mark [11]. Surprisingly, no examples of ubiquitin-binding domains have yet been identified in viral proteins. SUMO mediates protein-protein interactions via SIMs on receptor proteins [131]. SIMs have recently been identified in some viral proteins and these may play roles in nucleating viral structures, in promoting transcriptional activation, and in targeting SUMOylated cellular substrates for degradation. Given the extensive SUMOylation that takes place at DNA damage sites, it is likely that the SUMO mark on DDR proteins is also exploited by viruses for manipulation of the cellular response.
One of the major challenges in the study of ubiquitination is to identify substrates. Viral proteins have historically revealed many key cellular regulators and have defined important protein interactions. Proteomic studies with viral proteins will help to define protein interaction networks involved in the cellular DNA damage response and how they are perturbed during virus infection and transformation. The molecular surfaces that mediate protein-protein interactions between virus and host may also be attractive targets for development of small inhibitors as novel antiviral therapies.
Acknowledgments
We apologize to groups whose primary research papers were not cited due to space constraints or inadvertent oversight. We thank past and present members of the Weitzman lab for their helpful discussions and comments on the manuscript. MSC is supported by a predoctoral Ruth L. Kirschstein National Research Service Award (NIH/NCI T32 CA009523) and by a gift from the H.A. & Mary K. Chapman Charitable Trust. Work on viruses and DNA repair in the Weitzman lab has been supported by grants from the National Institutes of Health (AI067952, CA097093 and AI051686) and a Pioneer Developmental Chair from the Salk Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Huen MS, Chen J. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 2008;18:8–16. doi: 10.1038/cr.2007.109. [DOI] [PubMed] [Google Scholar]
- 6.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15:119–26. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 8.Al-Hakim A, Escribano-Diaz C, Landry MC, O’Donnell L, Panier S, Szilard RK, Durocher D. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 2010;9:1229–40. doi: 10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–89. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 10.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–26. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–71. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen ZJ, Sun LJ. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 14.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 15.Mohammad DH, Yaffe MB. 14–3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–17. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–40. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 18.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–14. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 20.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 21.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viswanathan K, Fruh K, DeFilippis V. Viral hijacking of the host ubiquitin system to evade interferon responses. Curr Opin Microbiol. 2010;13:517–23. doi: 10.1016/j.mib.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isaacson MK, Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–70. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11:527–34. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 25.Blanchette P, Branton PE. Manipulation of the ubiquitin-proteasome pathway by small DNA tumor viruses. Virology. 2008 doi: 10.1016/j.virol.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Banks L, Pim D, Thomas M. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem Sci. 2003;28:452–9. doi: 10.1016/S0968-0004(03)00141-5. [DOI] [PubMed] [Google Scholar]
- 27.Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76:841–50. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 29.Barry M, Fruh K. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci STKE. 2006;2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- 30.Harada JN, Shevchenko A, Pallas DC, Berk AJ. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J Virol. 2002;76:9194–206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Querido E, et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–17. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchette P, et al. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol Cell Biol. 2004;24:9619–29. doi: 10.1128/MCB.24.21.9619-9629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blackford AN, Grand RJ. Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation. J Virol. 2009;83:4000–12. doi: 10.1128/JVI.02417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng CY, Gilson T, Dallaire F, Ketner G, Branton PE, Blanchette P. The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity. J Virol. 2011;85:765–75. doi: 10.1128/JVI.01890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackford AN, et al. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc Natl Acad Sci U S A. 2010;107:12251–6. doi: 10.1073/pnas.0914605107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–95. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–95. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–52. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 40.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–20. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz RA, Lakdawala SS, Eshleman HD, Russell MR, Carson CT, Weitzman MD. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J Virol. 2008;82:9043–55. doi: 10.1128/JVI.00925-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Shevchenko A, Berk AJ. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol. 2005;79:14004–16. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo JL, Berk AJ. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J Virol. 2007;81:575–87. doi: 10.1128/JVI.01725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forrester NA, et al. Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection. J Virol. 2011;85:2201–11. doi: 10.1128/JVI.01748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathew SS, Bridge E. The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology. 2007;365:346–55. doi: 10.1016/j.virol.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 46.Lakdawala SS, Schwartz RA, Ferenchak K, Carson CT, McSharry BP, Wilkinson GW, Weitzman MD. Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J Virol. 2008;82:8362–72. doi: 10.1128/JVI.00900-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans JD, Hearing P. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J Virol. 2005;79:6207–15. doi: 10.1128/JVI.79.10.6207-6215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carson CT, et al. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 2009;28:652–62. doi: 10.1038/emboj.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X, Madden-Fuentes RJ, Lou BX, Pipas JM, Gerhardt J, Rigell CJ, Fanning E. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in Simian virus 40-infected primate cells. J Virol. 2008;82:5316–28. doi: 10.1128/JVI.02677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gregory DA, Bachenheimer SL. Characterization of mre11 loss following HSV-1 infection. Virology. 2008;373:124–36. doi: 10.1016/j.virol.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ. Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakasai R, Tibbetts R. RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage. J Biol Chem. 2008;283:13549–55. doi: 10.1074/jbc.M710197200. [DOI] [PubMed] [Google Scholar]
- 53.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:20759–63. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lilley CE, et al. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 2010;29:943–55. doi: 10.1038/emboj.2009.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lilley CE, Chaurushiya MS, Boutell C, Everett RD, Weitzman MD. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog. 2011 doi: 10.1371/journal.ppat.1002084. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lieberman PM. Chromatin regulation of virus infection. Trends Microbiol. 2006;14:132–40. doi: 10.1016/j.tim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9:930–6. doi: 10.4161/cc.9.5.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 59.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 60.Ikura T, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–40. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–7. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Awasthi S, et al. A human T-cell lymphotropic virus type 1 enhancer of Myc transforming potential stabilizes Myc-TIP60 transcriptional interactions. Mol Cell Biol. 2005;25:6178–98. doi: 10.1128/MCB.25.14.6178-6198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–66. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 64.Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38:700–11. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–81. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shanbhag NM, Greenberg RA. Neighborly DISCourse: DNA double strand breaks silence transcription. Cell Cycle. 2010;9:3635–6. doi: 10.4161/cc.9.18.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell. 2010;18:371–84. doi: 10.1016/j.devcel.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ginjala V, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage induced H2A ubiquitination and repair. Mol Cell Biol. 2011 doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson RL, Sawtell NM. Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J Virol. 2006;80:10919–30. doi: 10.1128/JVI.01253-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai W, Astor TL, Liptak LM, Cho C, Coen DM, Schaffer PA. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–12. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Preston CM, Nicholl MJ. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–13. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samaniego LA, Neiderhiser L, DeLuca NA. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–20. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Everett RD, Parsy ML, Orr A. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J Virol. 2009;83:4963–77. doi: 10.1128/JVI.02593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwiatkowski DL, Thompson HW, Bloom DC. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol. 2009;83:8173–81. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weiden MD, Ginsberg HS. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc Natl Acad Sci U S A. 1994;91:153–7. doi: 10.1073/pnas.91.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karen KA, Hoey PJ, Young CS, Hearing P. Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection. J Virol. 2009;83:4565–73. doi: 10.1128/JVI.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–12. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 79.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J Virol. 2007;81:7034–40. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muylaert I, Elias P. Knockdown of DNA ligase IV/XRCC4 by RNA interference inhibits herpes simplex virus type I DNA replication. J Biol Chem. 2007;282:10865–72. doi: 10.1074/jbc.M611834200. [DOI] [PubMed] [Google Scholar]
- 81.Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–7. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jackson SA, DeLuca NA. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc Natl Acad Sci U S A. 2003;100:7871–6. doi: 10.1073/pnas.1230643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeCaprio JA. How the Rb tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology. 2009;384:274–84. doi: 10.1016/j.virol.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 84.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:18159–64. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gonzalez SL, Stremlau M, He X, Basile JR, Munger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75:7583–91. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, Harper JW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81:9737–47. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–47. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levine AJ. The common mechanisms of transformation by the small DNA tumor viruses: The inactivation of tumor suppressor gene products: p53. Virology. 2009;384:285–93. doi: 10.1016/j.virol.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 89.Soria C, Estermann FE, Espantman KC, O’Shea CC. Heterochromatin silencing of p53 target genes by a small viral protein. Nature. 2010;466:1076–81. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bassermann F, Pagano M. Dissecting the role of ubiquitylation in the DNA damage response checkpoint in G2. Cell Death Differ. 2010;17:78–85. doi: 10.1038/cdd.2009.104. [DOI] [PubMed] [Google Scholar]
- 91.Heilman DW, Green MR, Teodoro JG. The anaphase promoting complex: a critical target for viral proteins and anti-cancer drugs. Cell Cycle. 2005;4:560–3. [PubMed] [Google Scholar]
- 92.Kornitzer D, Sharf R, Kleinberger T. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J Cell Biol. 2001;154:331–44. doi: 10.1083/jcb.200104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tran K, Mahr JA, Choi J, Teodoro JG, Green MR, Spector DH. Accumulation of substrates of the anaphase-promoting complex (APC) during human cytomegalovirus infection is associated with the phosphorylation of Cdh1 and the dissociation and relocalization of APC subunits. J Virol. 2008;82:529–37. doi: 10.1128/JVI.02010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tran K, Kamil JP, Coen DM, Spector DH. Inactivation and disassembly of the anaphase-promoting complex during human cytomegalovirus infection is associated with degradation of the APC5 and APC4 subunits and does not require UL97-mediated phosphorylation of Cdh1. J Virol. 2010;84:10832–43. doi: 10.1128/JVI.01260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim S, et al. HBV X protein targets hBubR1, which induces dysregulation of the mitotic checkpoint. Oncogene. 2008;27:3457–64. doi: 10.1038/sj.onc.1210998. [DOI] [PubMed] [Google Scholar]
- 96.Mo M, Fleming SB, Mercer AA. Cell cycle deregulation by a poxvirus partial mimic of anaphase-promoting complex subunit 11. Proc Natl Acad Sci U S A. 2009;106:19527–32. doi: 10.1073/pnas.0905893106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qiao X, Zhang L, Gamper AM, Fujita T, Wan Y. APC/C-Cdh1: from cell cycle to cellular differentiation and genomic integrity. Cell Cycle. 2010;9:3904–12. doi: 10.4161/cc.9.19.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–70. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Casey L, Wen X, de Noronha CM. The functions of the HIV1 protein Vpr and its action through the DCAF1.DDB1.Cullin4 ubiquitin ligase. Cytokine. 2010;51:1–9. doi: 10.1016/j.cyto.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.DeHart JL, Zimmerman ES, Ardon O, Monteiro-Filho CM, Arganaraz ER, Planelles V. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol J. 2007;4:57. doi: 10.1186/1743-422X-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dehart JL, Planelles V. Human immunodeficiency virus type 1 Vpr links proteasomal degradation and checkpoint activation. J Virol. 2008;82:1066–72. doi: 10.1128/JVI.01628-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang X, Pickering MT, Cho NH, Chang H, Volkert MR, Kowalik TF, Jung JU. Deregulation of DNA damage signal transduction by herpesvirus latency-associated M2. J Virol. 2006;80:5862–74. doi: 10.1128/JVI.02732-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–11. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Bie P, Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakada S, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–6. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 107.Canning M, Boutell C, Parkinson J, Everett RD. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J Biol Chem. 2004;279:38160–8. doi: 10.1074/jbc.M402885200. [DOI] [PubMed] [Google Scholar]
- 108.Boutell C, Canning M, Orr A, Everett RD. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J Virol. 2005;79:12342–54. doi: 10.1128/JVI.79.19.12342-12354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, Frappier L. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278:29987–94. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 110.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lindner HA. Deubiquitination in virus infection. Virology. 2007;362:245–56. doi: 10.1016/j.virol.2006.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19:547–57. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 113.Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J Virol. 2005;79:15582–5. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol. 2010;84:4026–40. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tang JB, Greenberg RA. Connecting the Dots: Interplay Between Ubiquitylation and SUMOylation at DNA Double Strand Breaks. Genes Cancer. 2010;1:787–796. doi: 10.1177/1947601910382774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–9. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 118.Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137–43. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hofmann H, Floss S, Stamminger T. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol. 2000;74:2510–24. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Adamson AL, Kenney S. Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol. 2001;75:2388–99. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chang LK, Liu ST, Kuo CW, Wang WH, Chuang JY, Bianchi E, Hong YR. Enhancement of transactivation activity of Rta of Epstein-Barr virus by RanBPM. J Mol Biol. 2008;379:231–42. doi: 10.1016/j.jmb.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 122.Endter C, Kzhyshkowska J, Stauber R, Dobner T. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc Natl Acad Sci U S A. 2001;98:11312–7. doi: 10.1073/pnas.191361798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chang PC, et al. Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J Biol Chem. 2010;285:5266–73. doi: 10.1074/jbc.M109.088088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pennella MA, Liu Y, Woo JL, Kim CA, Berk AJ. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J Virol. 2010;84:12210–25. doi: 10.1128/JVI.01442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Heideker J, Perry JJ, Boddy MN. Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair (Amst) 2009;8:517–24. doi: 10.1016/j.dnarep.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–12. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cherubini G, et al. The FANC pathway is activated by adenovirus infection and promotes viral replication-dependent recombination. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Boichuk S, Hu L, Hein J, Gjoerup OV. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. J Virol. 2010;84:8007–20. doi: 10.1128/JVI.00334-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Spardy N, Duensing A, Charles D, Haines N, Nakahara T, Lambert PF, Duensing S. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J Virol. 2007;81:13265–70. doi: 10.1128/JVI.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–8. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]