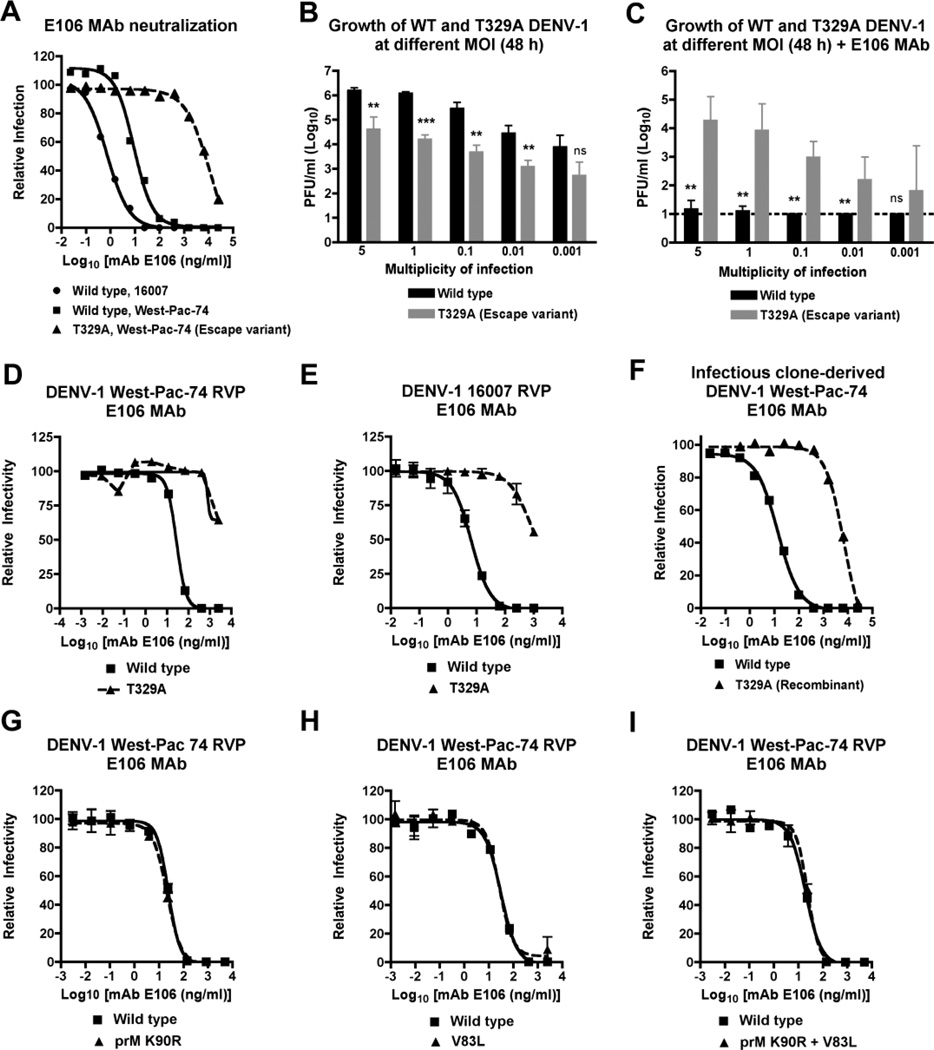
Figure 1. Phenotype of the DENV-1 T329A neutralization escape variant.
A. Neutralization curves with DENV1-E106 and the parent 16007 DENV-1 strain (genotype 2), the heterologous West-Pac-74 DENV-1 strain (genotype 4), and the T329A West-Pac-74 DENV-1 strain derived directly from passage in cell culture. The data are representative of three independent experiments performed in duplicate on BHK21-15 cells. B–C. Yield of wild type and T329A West-Pac-74 DENV-1 from supernatants of BHK21-15 cells after infection at different input MOI in the (B) absence or (C) presence of DENV1-E106. The data are the average of three independent experiments performed in duplicate and asterisks indicate differences that are statistically significant. Dashed lines indicate the limit of detection of the assay. D–E. Confirmation of resistant phenotype with wild type and mutant DENV-1 RVP. Genetically engineered DENV-1 RVP displaying the structural proteins of (D) West-Pac-74 or (E) 16007 with or without a single amino acid substitution (T329A) were incubated with increasing concentrations of DENV1-E106. F. Neutralization curves of DENV1-E106 with wild type or T329A West-Pac-74 DENV-1 strains derived by site-directed substitution into an infectious cDNA clone. The data are representative of two independent experiments performed in duplicate on BHK21-15 cells. G–I. Genetically engineered DENV-1 RVP (strain West-Pac-74) with single mutations (G) K90R in prM or (H) V83L in E or both mutations (I) K90R + V83L were incubated with increasing concentrations of DENV1-E106 and analyzed by flow cytometry. For D, E, and G–I, representative data of experiments performed on Raji-DCSIGNR cells is shown. Error bars represent the standard error of the mean of duplicate infections.