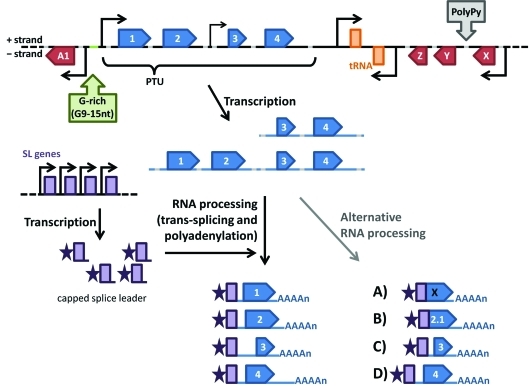
Figure 3.
Gene expression in trypanosomatids. Large clusters of unrelated genes (arrow boxes) are organized as polycistronic transcription units (PTUs) that are separated by divergent or convergent strand-switch regions. RNA Pol II transcription start sites (TSS) are usually located upstream of the first gene of the PTU (Martínez-Cavillo et al., 2004) or can be located as an internal TSS (Kolev et al., 2010). At the TSS (large bent arrow), the histone variants H2AZ and H2BV (Siegel et al., 2009), modified histones [K9/K14 acetylated and K4 tri-methylated histone (Respuela et al., 2008; Thomas et al., 2009; Wright et al., 2010) and K10 acetylated histone H4 (Siegel et al., 2009)], bromodomain factor BDF3 (Siegel et al., 2009) and transcription factors TRF4 and SNAP50 (Thomas et al., 2099) are frequently associated, with a few of these chromatin modifications also detected at internal TSS (small bent arrow) (Siegel et al., 2009; Wright et al., 2010). The polycistronic RNAs (pre-mRNAs) are individualized in monocistronic mRNAs after the addition of a capped splice leader RNA through a trans-splicing reaction coupled to polyadenylation. These processing reactions are guided by polypyrimidine tracts (PolyPy) that are present in every intergenic region. Mature mRNAs are exported to the cytoplasm where their stability and translation efficiencies are largely dependent on cis-acting elements present in their untranslated region (UTR) (Araujo et al., 2011). Transcriptomic analyses also showed that polycistronic pre-mRNAs can suffer alternative RNA processing that may result in changes in the initiator AUG, thereby altering protein translation (A), targeting and/or function (B). Alternative splicing and polyadenylation can also result in the inclusion/exclusion of regulatory elements present in the 5′ UTRs (C) or 3′ UTRs (D), thereby altering gene expression (Kolev et al., 2010; Nilsson et al., 2010; Siegel et al., 2010).